Control of fungal pathogens is mainly addressed by the use of chemically synthesized fungicides which result in environmental pollution, developing resistance after prolonged use. In this context, endophytes have been recognized as potential biocontrollers, and also as a promising source of antifungal metabolites. Therefore, as part of our research on phytopathogen controllers, 355 fungal endophytes were isolated from Protium heptaphyllum and Trattinnickia rhoifolia (Burseraceae), both ethnobotanically important tree species that produce secondary metabolites of agronomic and industrial interest. Endophytes were tested by in vitro dual culture against Fusarium oxysporum, a phytopathogen of agronomic importance. Five endophytes exerted at least 40% inhibition on F. oxysporum growth. Ethyl acetate (EtOAc) extracts were obtained from the most active antagonistic fungi, after growing them in three different liquid media. The extracts were tested against a conidial suspension of F. oxysporum by direct bioautography. Two extracts derived from fungi identified as Chaetomium globosum, F211_UMNG and Meyerozima sp. F281_UMNG showed inhibition of pathogen growth. Isolate C. globosum, F211_UMNG was selected for a chemical analysis by RP-HPLC-DAD-ESI-MS and antifungal molecules such as cladosporin, chaetoatrosin A and chaetoviridin A were annotated and identified based on their MS data.
El control de patógenos fúngicos se basa principalmente en el uso de fungicidas de síntesis química, los que pueden dar lugar a la contaminación del medio ambiente y el desarrollo de resistencia después de un uso prolongado. En este contexto, los endófitos han sido reconocidos como potenciales biocontroladores y también como fuentes prometedoras de metabolitos secundarios antifúngicos. En el marco de nuestra investigación sobre controladores de fitopatógenos, se aislaron 355 hongos endófitos de Protium heptaphyllum y Trattinnickia rhoifolia (Burseraceae), especies arbóreas de valor etnobotánico que producen metabolitos secundarios de interés agronómico e industrial. Los endófitos fueron evaluados in vitro en cultivos duales frente a Fusarium oxysporum, un fitopatógeno de importancia agronómica. Cinco endófitos mostraron al menos un 40% de inhibición en el crecimiento de F. oxysporum. Una vez determinados los hongos más activos, estos se cultivaron en 3 medios líquidos diferentes y a partir de ellos se preparó una serie de extractos solubles en acetato de etilo. Los extractos fueron probados contra una suspensión de conidios de F. oxysporum por bioautografía directa. Dos extractos derivados de los hongos identificados como Chaetomium globosum (F211_UMNG) y Meyerozima sp. (F281_UMNG) mostraron inhibición del crecimiento del patógeno. En el extracto derivado del hongo C. globosum se anotaron e identificaron los compuestos antifúngicos cladosporina, chaetoatrosina A y chaetoviridina A mediante el análisis por RP-HPLC-DAD-ESI-MS.
Using chemically synthesized fungicides has been the first line strategy to control phytopathogenic fungi31. However, secondary effects, such as environmental pollution and resistance development due to the use of these products, has led to a growing reluctance to use hazardous fungicides in agriculture. Thus, an enhanced trend in searching new control strategies involving environment-friendly alternatives in the management of plant pathogens has arisen20. In the search for such control strategies, naturally-occurring chemical entities have become potential alternatives for the industry to replace synthetic products23. In this context, microorganisms constitute a rich source of compounds with useful properties48 for several applications in the agrochemical and pharmaceutical industries11,23.
For several decades, the interaction between fungal endophytes and their hosts has attracted the researchers’ attention, mainly because of the advantageous characteristics they confer to their host. Among these characteristics we can mention enhanced stress tolerance, plant growth factor production, herbivore repellency and protection against pathogens18. The latter characteristic is partly due to the fact that endophytes compete with other microorganisms for a specific niche, which could be achieved by the production of antibiotic-like secondary metabolites, along with other strategies4. As a consequence of their repellent properties, endophytes have been proposed as biocontrollers and as a promising source of antifungal metabolites against phytopathogens of agronomic importance18.
Based on our ongoing search for biologically active secondary metabolites from endophytic fungi, the objective of this work was to explore the diversity of endophytes isolated from Protium heptaphyllum and Trattinnickia rhoifolia (Burseraceae) form Casanare, Colombia. These tree species, have been traditionally used by indigenous communities to treat several ailments12, and their complex chemical repertory has provided useful compounds having industrial, pharmaceutical and agronomic potential38,43. Furthermore, endophytes have been isolated from a species of the Burseraceae family, such as Muscodor yucatensis25, with potential for controlling phytopathogens. Therefore, the aim of this work was to test in vitro the abilities of endophytes to inhibit the mycelial growth of Fusarium oxysporum, by metabolite production. F. oxysporum is a pathogen of many plant species that represent a major threat for the production of several agriculturally important crops, such as banana, carnation, chickpeas, dates, lentils, tomato, and others27. The active component or components, responsible for the antifungal activity were partially characterized following a bioassay-guided fractionation test of the liquid culture-derived crude extract from the most antagonistic endophyte, to be incorporated in the future to control management programs for plant pathogen F. oxysporum.
MethodsRecovery of endophytes and isolationA total of two individuals from P. heptaphyllum and two from T. rhoifolia were collected in the foothill of the west Colombian Andes mountains in Aguazul, Casanare, Colombia (N 05°13′47.89″, W 072°30′31.38″), a transition ecosystem between the savanna and the high Andean ecosystems. Botanical specimens of P. heptaphyllum (Aubl.) Marchand (COL573961) and T. rhoifolia (Aubl.) Marchand (COL573962) were deposited in the Colombian National Herbarium. From each tree, the plant material (from higher, medium and lower strata) was sampled in order to collect representative isolates from all the plants. Five leaves per level were collected in a total of 60 leaflets that were bagged in sealed bags and stored in dark conditions for 24h at room temperature (ca. 26°C). Petioles were then removed and the complete leaves were vigorously washed with distilled sterile water and Tween 20 (0.01%), then submerged in 70% aqueous ethanol (1min), then in 1% sodium hypochlorite (3min), and then rinsed three times with sterilized distilled water. Leaves were then imprinted on Potato Dextrose Agar (PDA, Oxoid, UK) for verifying the disinfection of all epiphytic microorganisms.
Each leaf was sectioned into 2mm2 pieces, and 5 randomly-chosen pieces from each leaf were seeded in Petri dishes (90mm×15mm) containing water agar (agar 1.5%) (WA), 1/10 PDA (PDA at a 10th of the recommended concentration) or PDA, and then incubated at 26°C. Hyphae tips emerging from the leaf pieces were collected for three weeks, and sub-cultured on PDA at 26°C in the dark. Axenic cultures were established and, when a specific fungus sporulated, a monosporic culture was established. Those fungi that never sporulated were kept for a hyphal tip culture.
Identification of endophytesFungal populations were identified on the basis of cultural characteristics and morphology of fruiting bodies and spores3,14,21. Fungi were identified up to the genus level by observing the presence of conidial mycelium, spore mass color, distinctive reverse colony color, production of diffusible pigments, and spore morphology3. Cultures that repetitively failed to sporulate on different media were recorded as mycelia sterilia.
Additionally, those endophytes that inhibited F. oxysporum growth above 40% were identified by amplification of the nuclear ribosomal internal transcribed spacer (ITS) region, using the primers ITS1 (5′-TCC GTA GGT GAA CCT GCG G-3′) and ITS 4(5′-TCC TCC GCT TAT TGA TAT GC-3′)46. Amplicons were sequenced with the same primers bidirectionally a single time in Macrogen Inc. (Korea), and the resulting sequences were aligned and edited in BioEdit v7.2.513. The sequences were confronted with those in GeneBank database (http://www.ncbi.nlm.nih.gov), using BLASTN 2.2.2828. The closest match was selected and aligned using ClustalW26. For the phylogenetic analysis, tree constructions were done with the MEGA 6.0 program package41 using the neighbor-joining method37. Bootstrap analysis was done using 1000-times resampled data. The resulting sequences were deposited in the GenBank.
Ethyl acetate (EtOAc) extractionFungi selected for their inhibitory activity at in vitro conditions against F. oxysporum were reactivated in 500ml of Potato Dextrose Broth (PDB, Oxoid, UK), Sabouraud Broth (SAB, Oxoid, UK) and yeast extract sucrose media (YES)19, and cultured in an orbital shaker under constant agitation (100rpm) at 21°C for 7 days. After that period the culture was filtered using Whatman No. 1 qualitative filter paper, and mycelia were lyophilized. Separately, both mycelia and filtered media were mixed with EtOAc in a 1:3 proportion and incubated in an orbital shaker in constant agitation (100rpm) for 48h. The organic phase (EtOAc) was separated from the mycelia by vacuum filtration using Whatman No. 1 qualitative filter paper, and from filtered liquid media using a decantation funnel. The resulting extracts were concentrated by lyophilization.
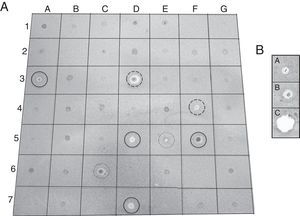
Antifungal assaysFungal endophytes and a phytopathogenic isolate (F. oxysporum G1 isolated from Physalis peruviana (Cape gooseberry) available in the collection of the phytopathology laboratory at Universidad Militar Nueva Granada) were cultured on PDA at 26°C for 5 days at 26°C in the dark. In order to evaluate the possible effect of each endophyte on phytopathogen growth, dual cultures were settled and each isolate was challenged with F. oxysporum G1. Thus, a plug (3mm diameter), which was obtained from the colonial actively growing edge of the endophyte to be tested, was seeded on PDA, 10mm away from the edge of a Petri dish (90mm×10mm). At a spot distance (10mm) from the diametrally-opposed edge, a similar plug of F. oxysporum was seeded. Six days later, the effect of each endophyte on F. oxysporum growth was observed and F. oxysporum colony radial measurement and distance between colonies were recorded. As control, a plug of each organism was cultured alone. These experiments were replicated three times. Results were compared by the Tukey's HSD (honest significant difference) test.
The antifungal activity of the extracts was tested by direct TLC bioautographic detection9. Extracts and fractions from the selected endophytes (Table 1) were diluted in ethanol (HPLC grade) and 30μg were seeded in a single spot on a TLC Aluminum silica gel 60 Sheet 20cm×20cm (Sigma–Aldrich). Then the silica sheet was sprayed with a 1×106conidia/ml conidial suspension of F. oxysporum until the whole sheet was covered. The assays were incubated in a humid chamber for 3 days, and then F. oxysporum growth over the sheet was evaluated under UV-light.
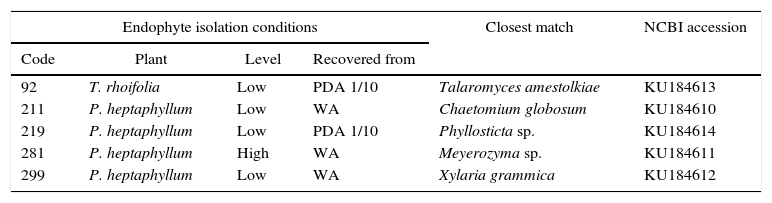
Isolation media and closest match in phylogenetic analysis by the neighbor joining method form the most active endophytes against F. oxysporum
| Endophyte isolation conditions | Closest match | NCBI accession | |||
|---|---|---|---|---|---|
| Code | Plant | Level | Recovered from | ||
| 92 | T. rhoifolia | Low | PDA 1/10 | Talaromyces amestolkiae | KU184613 |
| 211 | P. heptaphyllum | Low | WA | Chaetomium globosum | KU184610 |
| 219 | P. heptaphyllum | Low | PDA 1/10 | Phyllosticta sp. | KU184614 |
| 281 | P. heptaphyllum | High | WA | Meyerozyma sp. | KU184611 |
| 299 | P. heptaphyllum | Low | WA | Xylaria grammica | KU184612 |
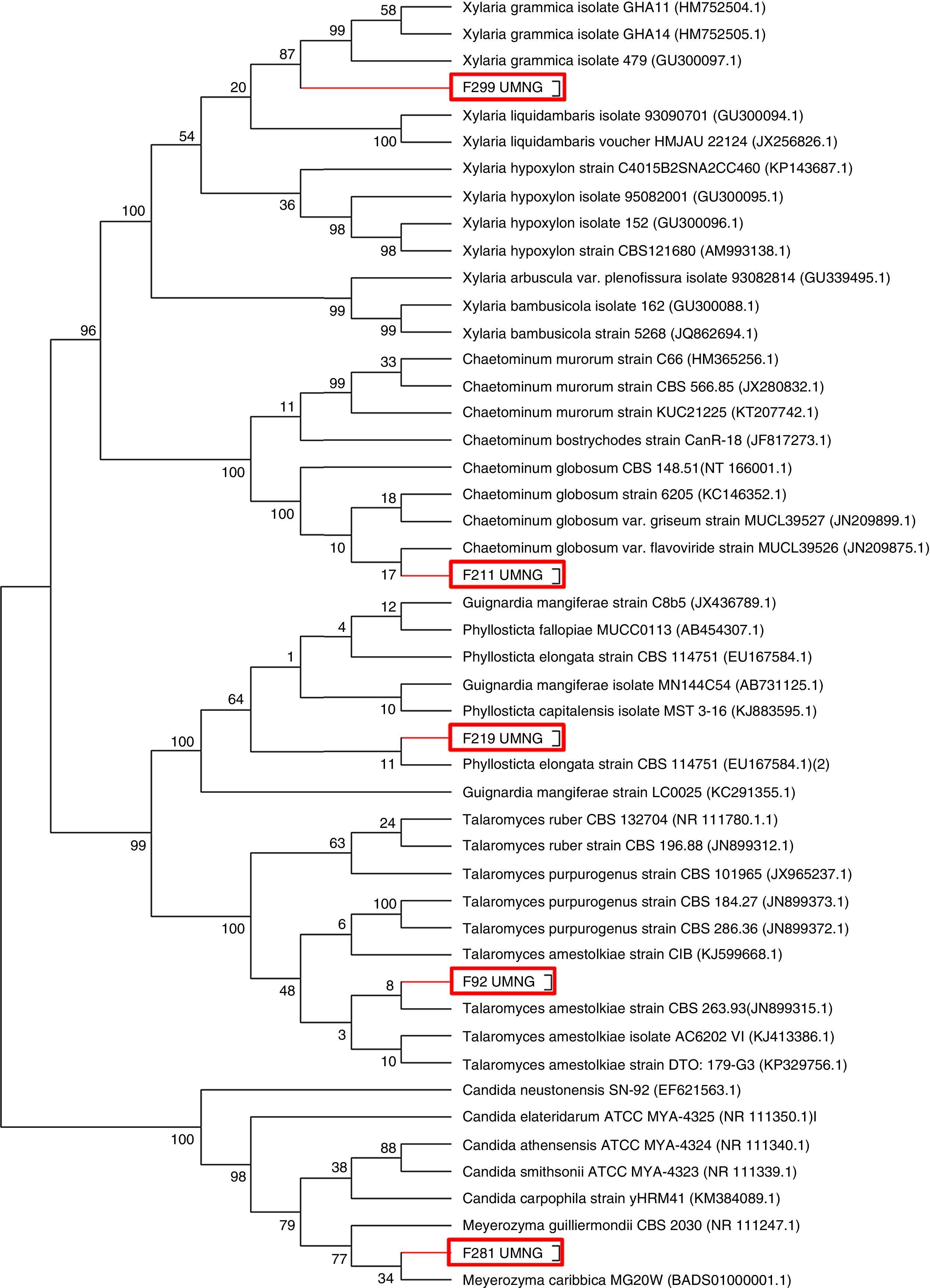
Species were defined when the node was supported with ≤90 (Fig. 1).
The most active extract was fractionated by preparative HPLC (Shimadzu prominence LC20AD), in gradient elution, using a Shimadzu Premier column C-18 (4.6mm×150mm, 5μm) at a flow rate of 2ml/min. The injection volume was 50μl. The mobile phases consisted in methanol (HPLC grade) (Phase A) and trifluoroacetic acid 0.005% (HPLC grade) (Phase B). Separation was carried out for 25min, in a FRC 10A Shimadzu fraction collector. A diode array detector (DAD) performed signal detection at 270nm. A total of 20 fractions were recovered and then concentrated by lyophilization.
LC-MS-based chemical analysisExtracts and fractions were characterized by Reverse Phase Liquid Chromatography with multi-wavelength UV-VIS detection (by a DAD) and coupled by electrospray to a mass spectrometry detector (RP-HPLC-DAD-ESI-MS) (Shimadzu Prominence LC/MS 8030). Analyses were performed on a Shimadzu prominence instrument, in gradient elution, using a Shimadzu Premier column C-18 (4.6mm×150mm, 5μm). Simultaneous monitoring was carried out at 270nm, at a flow rate of 0.6ml/min. The operating temperature was 30°C and the injection volume was 20μl. As mobile phase A 1% formic acid in distilled water (HPLC grade) was used, and acetonitrile (ACN) (HPLC grade) as mobile phase B; separation was performed for 33min. The mass spectrometry detector (MSD) consisted of an electrospray ionization (ESI) source and a triple quadrupole analyzer. The mass spectrometry method consisted of a scan in simultaneous positive and negative ionization with an acquisition time of 2–33min, a mass range of 50–2000m/z, a scan speed of 1667μ/s, an event time of 0.5s, nebulizer gas flow of 1.5l/min, 350°C interface temperature and DL, and 450°C block temperature. The drying gas flow rate was 9l/s. The analysis was monitored at wavelengths between 270 and 330nm. Annotation and identification of the major and minor metabolites in the extract was performed by mass spectrometry- based analysis, complemented with the analysis of reported metabolites.
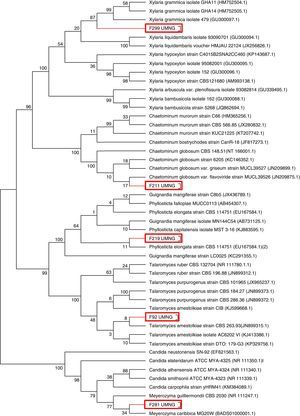
ResultsRecovery of endophytesA total of 577 endophytes were isolated from 900 cultured pieces of leaflets. A subtotal of 355 endophytes were selected after the elimination of redundant morphotypes derived from the same leaflet. The highest number of endophytes (n=236) was recovered from the lowest collection level for both species. The ITS region of the isolates was amplified and sequenced to determine the phylogenetic relationships among them (Fig. 1). A phylogenetic tree was constructed based on a 570bp sequences and isolates clustered as follows: endophyte F92_UMNG clustered with Talaromyces amestolkiae, F18_UMNG with Phyllosticta sp., F211_UMNG with Chaetomium globosum, F299_UMNG with Xylaria grammica, and F281_UMNG with Meyerozyma sp. (Table 1).
Dendrogram showing the phylogenetic relationship of fungal endophytes based on the ITS region. Phylogenies were inferred using the neighbor-joining analysis and trees generated in MEGA 6.0 software. Numbers at branch points indicate bootstrap values. The scale bars represent the estimated difference in nucleotide sequence. Red rectangles indicate the endophytes isolated in this work.
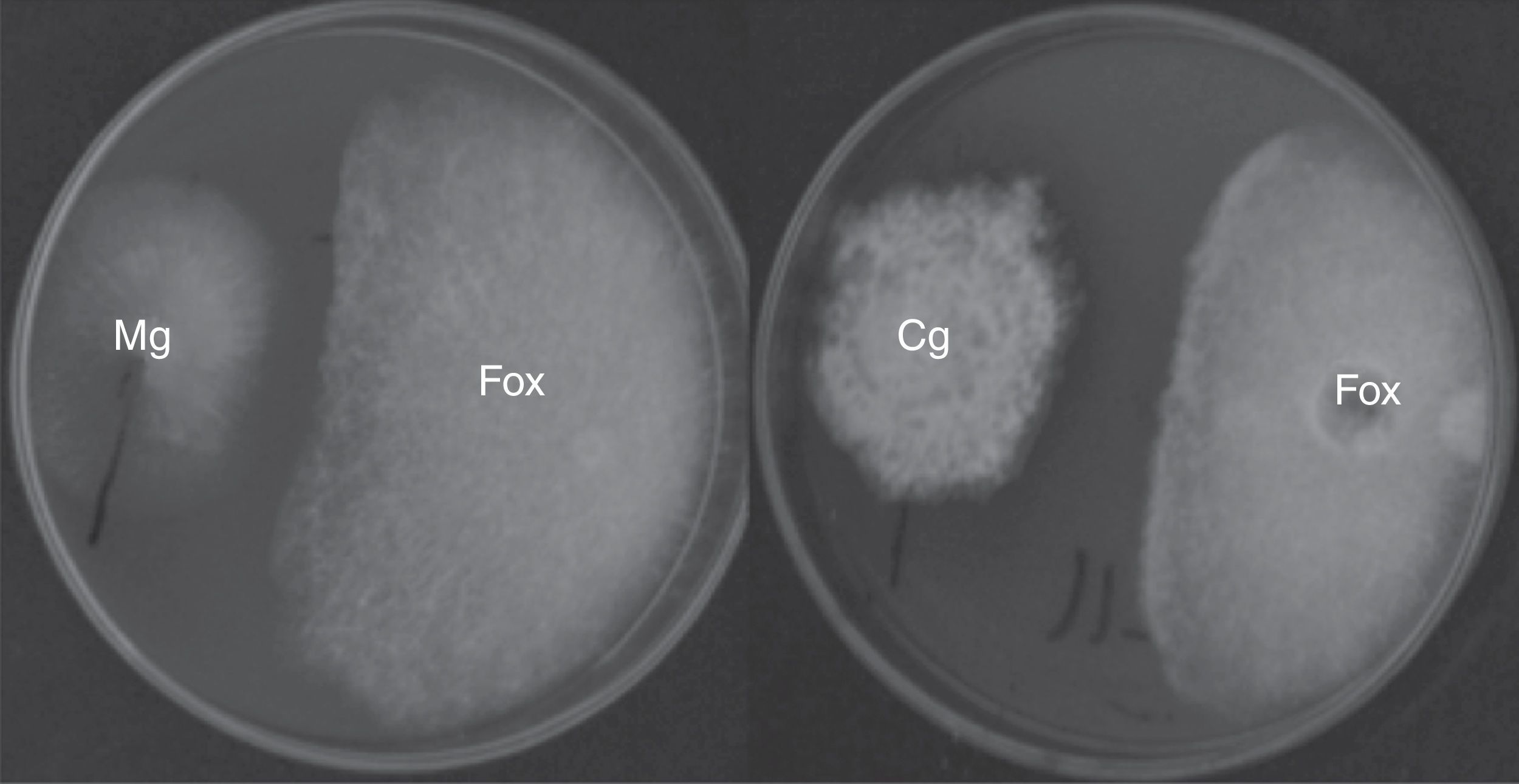
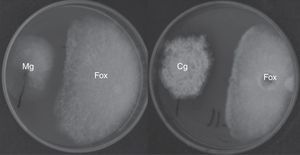
The antifungal ability of 355 fungal endophytes against F. oxysporum G1 was evaluated by the dual culture method. Five endophytes reduced the area of F. oxysporum growth by at least 40%, without colony contact (Fig. 2), which were grouped in a single group by the Tukey's HSD test.
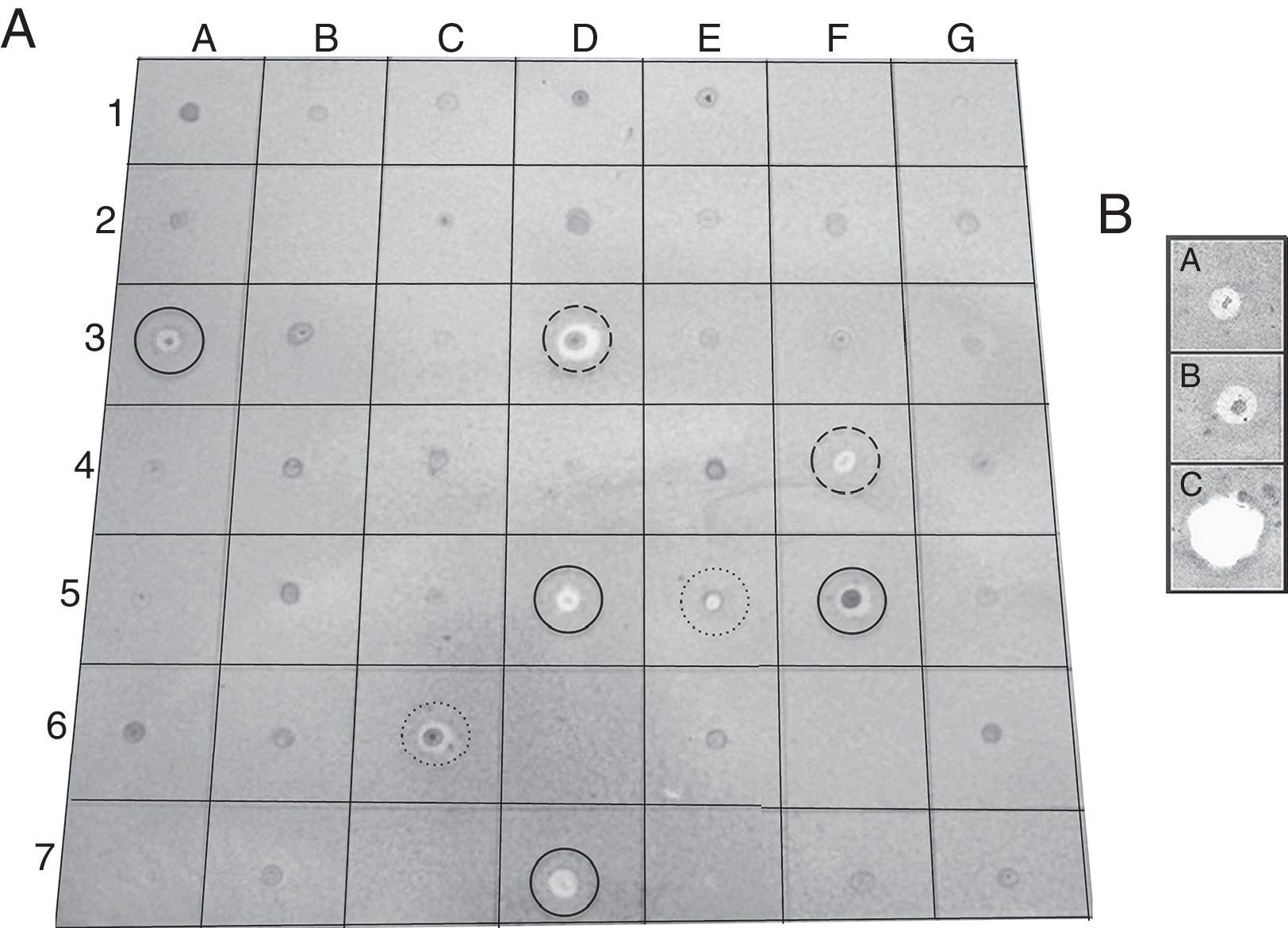
EtOAc-soluble extracts obtained from two isolates, F211_UMNG and F281_UMNG, cultured in YES, exerted inhibitory effect on F. oxysporum by direct bioautography (Fig. 3A). On comparing the inhibitory effect produced by these two isolates on F. oxysporum radial growth, it was found that isolate F211_UMNG (C. globosum) exerted a 64% in vitro inhibition of F. oxysporum colony growth arresting its growth producing with a distance between colonies of 12.5±0.6mm while F281_UMNG caused 45% inhibition showing 5.7±0.9mm between colonies (Figs. 2 and 3A).
(A) Direct bioautography of endophyte-derived EtOAc extracts against F. oxysporum conidia. Solid lines: Procloraz 40ng (control). Medium dashed line: Supernatant medium of EtOAc extract from C. globosum F211_UMNG. Highly dashed line: Supernatant medium of EtOAc extract from M. guilliermondii F281_UMNG. (B) Bioautography of F211_UMNG extract and most active fraction against F. oxysporum conidia. A: Fraction #14, B: Extract 211 INI, C: Sportak 40ng.
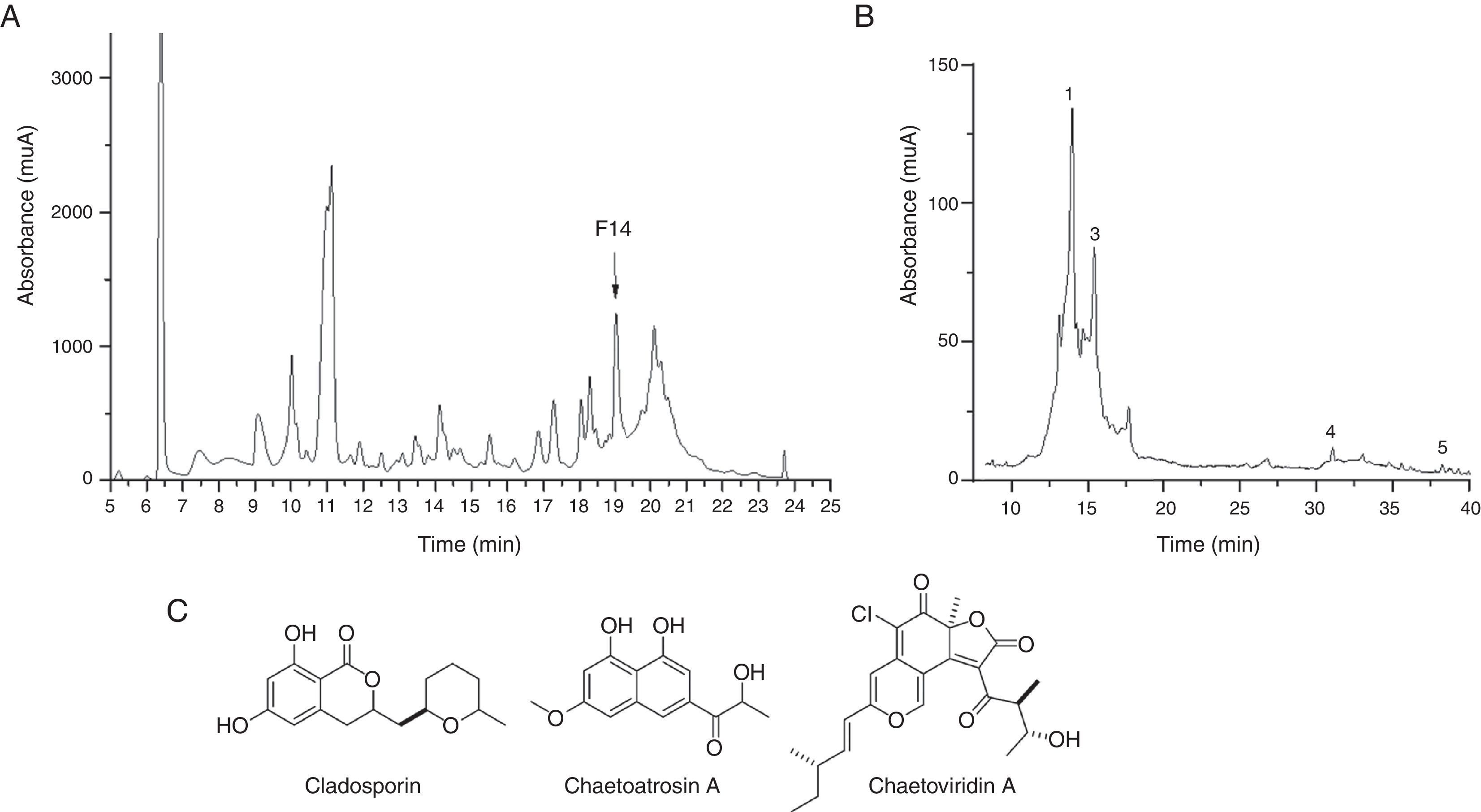
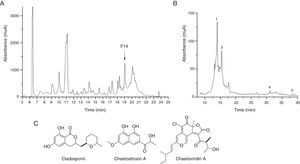
Since the extract from F211_UMNG isolate showed greater inhibitory activity against F. oxysporum, it was fractionated in order to determine the most active fractions. A total of 20 fractions were recovered (Fig. 4A) and were additionally tested by bioautography against a conidial suspension of F. oxysporum. Only fraction #14 (30μg) exerted inhibition of the fungus (Fig. 3B). Fraction #14 was analyzed by LC-MS, rendering a chromatogram that included two defined signals between min 12 and min 17 (peaks 1 and 3, Fig. 4B).
(A) Chromatographic profile of EtOAc extract from F211_UMNG in YES medium. F14+ arrow indicates fraction #14. (B) RP-HPLC-DAD chromatogram of fraction #14. Numbers indicate the annotated compounds by MS data (Table 2). (C) Structures of the identified compounds in the most active fraction from EtOAc-soluble extract of C. globosum F 211_UMNG.
From the recovered isolates, fungi like Alternaria sp., Aspergillus sp., Chaetomium sp., Epicoccum sp., Fusarium sp., Pestalotiopsis sp., Phomopsis sp., Xylaria sp., among others, were identified by their morphological traits and have been previously reported as common endophytes in other plants6–8,22,24,29,47,49. A high diversity of fungal species were also found in leaves and stems of Boswellia sacra (Burseraceae), being Alternaria and Aspergillus the most dominant genera, which were both also isolated in this work. However, Chaetomium was also found in a relative high proportion (26.3%) represented by two species, C. globosum and C. spirale10. Screening works in Boswellia serrata exhibited Myrothecium verrucaria and Phoma sp.40 as dominant endophytes, which were also isolated in our samples.
The 355 endophytes isolated in this work were evaluated by the dual culture method against F. oxysporum and only five endophytes showed inhibition against F. oxysporum presumably by metabolite production because they inhibited the extension on the colony without mycelial contact and reduced the area of the phytopathogen by at least 40% (Table 1, Fig. 2). Antagonistic endophytes were identified by amplification of a 570bp (ITS) region as C. globosum, Meyerozyma spp., Phyllosticta spp., T. amestolkiae, and X. grammic; such species have been reported as being endophytes and having antibiotic activity1,2,17,33,35,39,49.
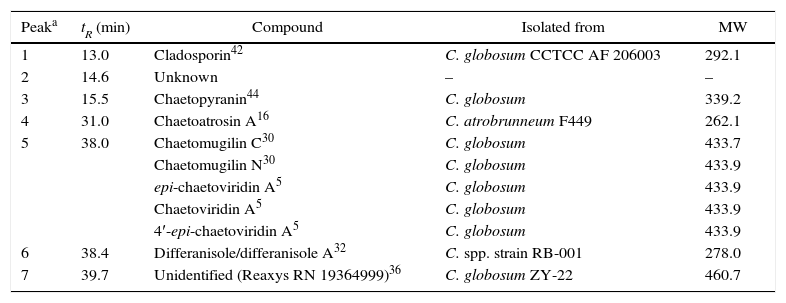
The extract from isolate C. globosum F211_UMNG, at 30μg, caused inhibit they inhibited the extension on the colony ion of F. oxysporum growth (Fig. 3A). Based on the available literature, in the Chaetomium genus, mostly in C. globosum, seven signals defined by mass spectrum analysis were found to have the same m/z value to that reported (Table 2). Nevertheless, five isomers previously reported in the Chaetomium genus matched the m/z value detected at 38min (peak 5, Fig. 4B) and, therefore they cannot be differentiated in accordance with the known MS limitations. Previous studies found that C. globosum synthetized several molecules such as chaetoglobosins, epipolythiodioxopiperazines, azaphilones, xanthones, anthraquinones, chromones, depsidones, terpenoids, and steroids, among others. These types of compounds have shown antitumor, cytotoxic, antimalarial, enzyme inhibitory, antibiotic, and other activities50. In the present study cladosporin, chaetoatrosin A and chaetoviridin A (Fig. 4C) were identified to be active against F. oxysporum, in the EtOAc extract of C. globosum F211_UMNG. These compounds were previously reported as having antifungal activity16,34,45. Chaetoatrosin A acts as an inhibitor of chitin synthase II, while chaetoviridin A inhibits the cholesteryl ester transfer protein (CETP)42. The action of cladosporin is no fully understood but it has been reported that it exhibited lysyl-tRNA synthetase inhibition in P. falciparum15 and that its mode of action is different to that affecting ß-tubuline assembly in mitosis45. The combination of the modes of action of the identified molecules might rationalize the observed growth inhibition of F. oxysporum in the in vitro and bioautography test.
Identified compounds isolated from other Chaetomium strains as constituents of fraction #14
| Peaka | tR (min) | Compound | Isolated from | MW |
|---|---|---|---|---|
| 1 | 13.0 | Cladosporin42 | C. globosum CCTCC AF 206003 | 292.1 |
| 2 | 14.6 | Unknown | – | – |
| 3 | 15.5 | Chaetopyranin44 | C. globosum | 339.2 |
| 4 | 31.0 | Chaetoatrosin A16 | C. atrobrunneum F449 | 262.1 |
| 5 | 38.0 | Chaetomugilin C30 | C. globosum | 433.7 |
| Chaetomugilin N30 | C. globosum | 433.9 | ||
| epi-chaetoviridin A5 | C. globosum | 433.9 | ||
| Chaetoviridin A5 | C. globosum | 433.9 | ||
| 4′-epi-chaetoviridin A5 | C. globosum | 433.9 | ||
| 6 | 38.4 | Differanisole/differanisole A32 | C. spp. strain RB-001 | 278.0 |
| 7 | 39.7 | Unidentified (Reaxys RN 19364999)36 | C. globosum ZY-22 | 460.7 |
In conclusion, five endophytes acting as antagonists of F. oxysporum under in vitro conditions were isolated and identified in the present study. Isolate C. globosum F211_UMNG-derived extract inhibits the growth of F. oxysporum, possibly by at least three molecules having different modes of action, implying its possible application in control schemes of F. oxysporum27. A confirmation of the results through in vivo testing is required, involving endophyte C. globosum and purifying the identified compounds for evaluating their ability in the control of the disease caused by F. oxysporum.
Ethical responsibilitiesProtection of human and animal subjectsThe authors declare that no experiments were performed on humans or animals for this study.
Confidentiality of dataThe authors declare that they have followed the protocols of their work center on the publication of patient data.
Right to privacy and informed consentThe authors declare that no patient data appear in this article.
FundingThe present work was financed by Vicerrectoría de Investigaciones at UMNG through the project INV-CIAS-1472.
Conflict of interestThe authors declare that they have no conflicts of interest.
Authors thank Universidad Militar Nueva Granada (UMNG) for the financial support. The present work is a product derived from the Project CIAS-1472 financed by Vicerrectoría de Investigaciones at UMNG.