In Argentina, information about epidemiology and environmental distribution of Cryptococcus is scarce. The city of Resistencia borders with Brazil and Paraguay where this fungus is endemic. All these supported the need to investigate the ecology of the genus and the epidemiology of cryptococcosis in this area.
AimsThe aim was to investigate the presence of species of Cryptococcus neoformans–Cryptococcus gattii complex and their genotypes in trees of the city of Resistencia.
MethodsOne hundred and five trees were sampled by swabbing technique. The isolates were identified using conventional and commercial methods and genotyped by PCR-RFLP (Restriction Fragment Length Polymorphism).
ResultsCryptococcus was found in 7 out of the total trees. 6 out of 7 Cryptococcus isolates were identified as C. neoformans and one as C. gattii. C. gattii was isolated from Grevillea robusta. C. neoformans strains were isolated from Tabebuia avellanedae and Peltophorum dubium. Genotyping showed that all C. neoformans belonged to the VNI type and C. gattii belonged to the VGI type.
ConclusionsThis represents the first study on the ecology of Cryptococcus spp. associated to trees from northeastern Argentina, and the first report describing Grevillea robusta as a host of members of this fungal genus. Another finding is the isolation of C. neoformans from Tabebuia avellanedae and Peltophorum dubium, both tree species native to northeastern Argentina.
En Argentina la información sobre la epidemiología y la distribución ambiental de Cryptococcus es escasa.. Resistencia es una ciudad que limita con Brasil y Paraguay, donde este hongo es endémico. Esto apoya la necesidad de investigar la ecología de este género y la epidemiología de la criptococosis en la región.
ObjetivosEl objetivo del presente estudio fue investigar la presencia de especies del complejo Cryptococcus neoformans – Cryptococcus gattii y sus genotipos en árboles de la ciudad de Resistencia, situada en el nordeste argentino.
MétodosMediante la técnica del hisopo se tomaron muestras de 105 árboles. Los aislamientos se identificaron utilizando métodos convencionales y comerciales, y se genotipificaron mediante la prueba PCR-RFLP (Restriction Fragment Length Polimorphism).
ResultadosSe aisló Cryptococcus en 7 árboles. Se identificaron 6 aislamientos como Cryptococcus neoformans y uno como Cryptococcus gattii. Este último se aisló de Grevillea robusta. Cryptococcus neoformans se aisló de Tabebuia avellanedae y Peltophorum dubium. La genotipificación mostró que todos los aislamientos de C. neoformans pertenecían al tipo molecular VNI, y C. gattii al tipo molecular VGI.
ConclusionesEl presente estudio es la primera investigación sobre la ecología del género Cryptococcus asociado a árboles del nordeste argentino, y la primera que describe Grevillea robusta como nicho ecológico de este género fúngico. Otro hallazgo es el aislamiento de C. neoformans de Tabebuia avellanedae y Peltophorum dubium, ambas especies de árboles originarias del nordeste argentino.
The name Cryptococcus refers to a haploid and heterothallic group of encapsulated yeasts classified within the phylum Basidiomycota. Its teleomorph is Filobasidiella, a filamentous fungus belonging to the class Tremellomycetes.5
This genus is characterized by having a polysaccharide capsule that is its main factor of virulence. There are about 70 species of Cryptococcus, most of which live in the soil and are not harmful to humans.15
As currently accepted, the Cryptococcus neoformans complex contains two species that cause cryptococcosis in both humans and animals, a disease that cause significant morbidity and mortality in immunocompromised and immunocompetent patients.27,38
The differentiation into two species is due to genetic differences, the composition of the capsular polysaccharide structure, the teleomorph, the biochemical properties, the geographical distribution, the reservoir and the immunological state of the host to whom it affects.17,19,29,30,38
The species and varieties belonging to the C. neoformans–C. gattii complex are C. gattii (serotypes B and C) and C. neoformans, with its two varieties, C. neoformans var. grubii (serotype A) and C. neoformans var. neoformans (serotype D). There is also a hybrid of these two varieties, serotype AD. Molecular methods have allowed the discrimination of eight main genetic types: VNI and VNII (C. neoformans var. grubii, serotype A), VNIII (C. neoformans serotype AD), VNIV (C. neoformans var. neoformans, serotype D), VGI, VGII, VGIII, and VGIV (C. gattii, serotypes B and C).3,31
Since the infection is acquired by inhalation of infective propagules present in the environment, it is important to study the habitat of this fungus.38,40
Either one or both species of Cryptococcus have been found in different families and genera of trees from around the world.1,2,16,26,35,41C. gattii had been considered to be restricted to areas of tropical and subtropical climate, although there is evidence that it has expanded its distribution.6,11,16,18,19,24,42
In Argentina, although the incidence of cryptococcosis has increased 40- to 50-fold from the AIDS pandemic, the knowledge of the environmental distribution of Cryptococcus is scarce and epidemiological information is fragmented, scanty and based only on the communications from hospitals assisting HIV-positive patients.7,32,33
Currently C. gattii is endemic in Latin America. In Brazil, cryptococcosis by C. gattii is prevalent in immunocompetent patients, and is the 8th cause of meningitis, the majority of the cases related to children below 14 years of age.39,43 Paraguay is among the Latin-American countries with high prevalence of this fungus, and outbreaks of infection with this agent have been reported in this country.8,24,25,43 North-eastern Argentina borders with Brazil and Paraguay. Resistencia city is located in this region and the lack of knowledge about the presence of this fungus in the environment supports the need to investigate the ecology of the genus and the epidemiology of cryptococcosis in this zone.
The aim of this study was to investigate the presence and genotypes of members of the Cryptococcus neoformans–Cryptococcus gattii species complex in trees of the city of Resistencia.
Materials and methodsStudy areaThe study was performed in the city of Resistencia (27°27′05″ S–58°59′12″ W), located in northeastern Argentina. The city is located 51m above sea level and is surrounded by rivers and lakes. The climate is subtropical with no dry season, with a mean annual rainfall of 1560mm. The mean annual temperature is 21°C, with extreme variations reaching 45°C in summer and 0°C in winter.37
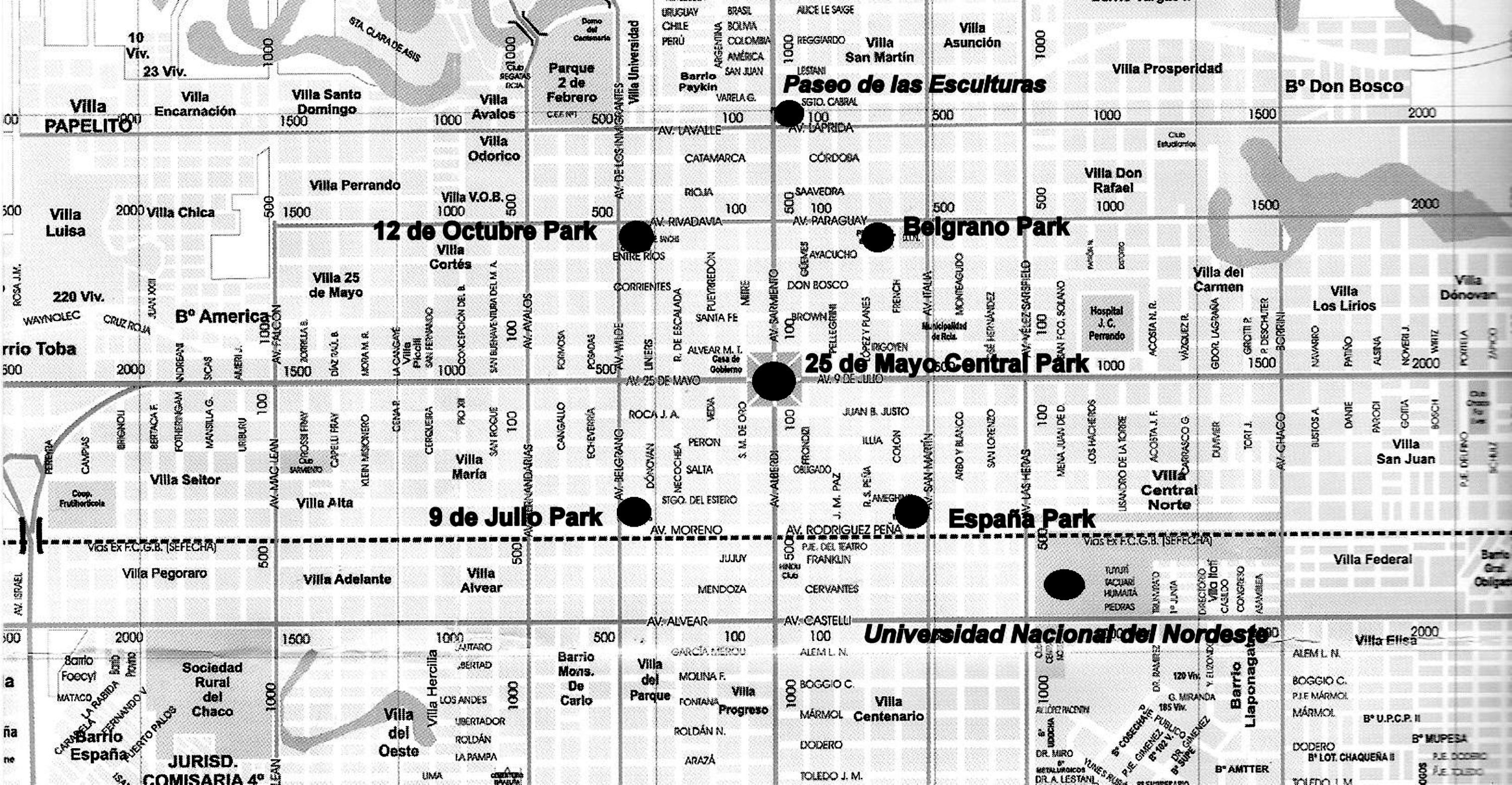
Samples were taken from five parks, as well as from the Universidad Nacional del Nordeste campus and the Paseo de las Esculturas, all spaces that are centrally located in downtown. The 25 de Mayo Central Park has 4ha, whereas the other four parks (Belgrano Park, España Park, 12 de Octubre Park, 9 de Julio Park) have only 1ha. The latter are equidistant, about 600m away from the central park. The campus is 1500m to the south of the central park, whereas the Paseo de las Esculturas is 800m to the northeast. The former has 5ha whereas the latter has 0.50ha (Fig. 1).
Swabbing techniqueSamples were collected by rubbing the inner of hollows or fissures of trees with a 5mm of diameter sterile cotton-tipped swab moistened in sterile saline solution (0.85% NaCl) supplemented with chloramphenicol (10mg/ml). Each swab was placed in a test tube with 3ml of the solution.21
IsolationWithout removing the swab, each tube was shaken manually for 5min. The swab was then removed allowing the solution to settle for 10min. The supernatant was diluted 1:10 in sterile saline solution. Then, 100μl of the supernatant and 100μl of the dilution were inoculated in separate plates of Pal medium34 supplemented with biphenyl (0.1%). The inoculated plates were incubated at 32°C.
Biochemical and physiological identification of the isolatesAfter 48h and up to 7 days, all brown colonies that grew on Pal medium were subcultured on Sabouraud medium. Cryptococcus isolates were identified according to the methodology of Kreger-van Rij20 and with the commercial API ID32 method (bioMérieux, Marcy l’Etoile-France). C. gattii was identified by culturing the isolates on canavanine-glycine-bromothymol blue medium,23 and by testing D-proline assimilation.10
Molecular analysisCryptococcus isolates were genotyped by URA5 gene RFLP (Restriction Fragment Length Polymorphism) analysis. DNA extraction was performed according to the procedures described by Bosco Borgeat et al.4 The PCR-RFLP reaction was performed as described by Meyer et al.31 The PCR products were double digested with Sau96I (Fermentas) and HhaI (Fermentas) and separated in 3% agarose gel (Biodynamics, Argentina) electrophoresis at 100V for 5h. RFLP patterns were assigned visually by comparing them with the patterns obtained from the reference strains (C. gattii: CBS 10078 VGI; CBS 10082 VGII; CBS 10081 VGIII; CBS 10101 VGIV. C. neoformans var. grubii: CBS 10085 VNI; CBS 10084 VNII. C. neoformans hybrid AD: CBS 10080 VNIII. C. neoformans var. neoformans: CBS 10079 VNIV).
ResultsOne hundred and five trees were sampled between March and December 2010. Cryptococcus was found in seven trees (6.7%).
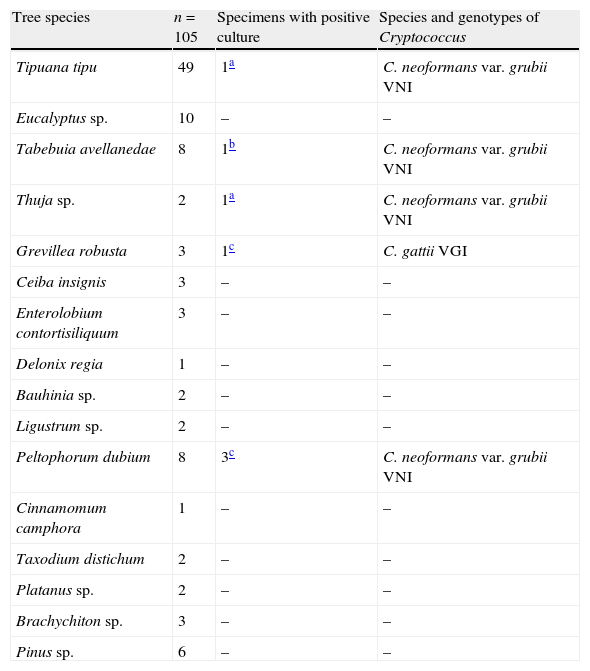
Six out of the seven specimens were identified as C. neoformans (85.7%) and one as C. gattii (14.3%). All isolates were collected from different trees. Table 1 shows the tree species sampled and which of them showed the presence of Cryptococcus.
Distribution of isolates and trees in the different parks sampled.
| Tree species | n=105 | Specimens with positive culture | Species and genotypes of Cryptococcus |
| Tipuana tipu | 49 | 1a | C. neoformans var. grubii VNI |
| Eucalyptus sp. | 10 | – | – |
| Tabebuia avellanedae | 8 | 1b | C. neoformans var. grubii VNI |
| Thuja sp. | 2 | 1a | C. neoformans var. grubii VNI |
| Grevillea robusta | 3 | 1c | C. gattii VGI |
| Ceiba insignis | 3 | – | – |
| Enterolobium contortisiliquum | 3 | – | – |
| Delonix regia | 1 | – | – |
| Bauhinia sp. | 2 | – | – |
| Ligustrum sp. | 2 | – | – |
| Peltophorum dubium | 8 | 3c | C. neoformans var. grubii VNI |
| Cinnamomum camphora | 1 | – | – |
| Taxodium distichum | 2 | – | – |
| Platanus sp. | 2 | – | – |
| Brachychiton sp. | 3 | – | – |
| Pinus sp. | 6 | – | – |
A second sampling was performed in positive trees obtaining the same RFLP genotypes.
Genotyping showed that all C. neoformans belonged to the variety grubii type VNI and that C. gattii belonged to the VGI type.
DiscussionCryptococcus gattii was first isolated by Ellis and Pfeiffer in 1990 from Eucalyptus trees in Australia and, for a long time, Eucalyptus spp. were pointed out as the ecological niche of C. gattii.12 However, the isolation of C. gattii from Eucalyptus spp. outside Australia was considered as rare.41 Our findings are consistent with the latter, since none of the Eucalyptus trees sampled showed the presence of Cryptococcus species. Affirming this, C. gattii has been found in 54 species of trees that are native to temperate, tropical and subtropical climates.41 In the present study, C. gattii was isolated only from an old specimen of Grevillea robusta. This is the first report that refers to this species as a host for this fungus. There are many specimens of this species it in the city of Resistencia and most of them are centenarians. The persistence of C. gattii in the same selected tree holes was confirmed by successive sampling. In agreement with other authors, this result shows that the ecological niche of C. gattii is not restricted to specific tree species.36,41
The natural habitat of C. neoformans has long been associated with nests of pigeons and other places with avian excreta accumulation.12,13,22,28 However, the presence of this species has been confirmed in holes of tree trunks with obvious degradation of lignin.40 In the present study, C. neoformans was found in Tabebuia avellanedae and Peltophorum dubium, both tree species native from Argentina. We found no references about these tree species as niche for any species of the genus Cryptococcus.
Tipuana tipu, a tree where C. neoformans was also found, is native from northwestern Argentina, in particular the Yungas and the subtropical forests of Bolivia. Because of its adaptability, Tipuana tipu has a wide distribution in Argentina, both by natural means and by the hand of man. In the city of Buenos Aires, it also has been reported harboring this yeast.36
The fact that all C. neoformans found belong to the VNI genotype is in agreement with reports from several countries indicating the VNI as the most frequent molecular type both as agent of cryptococcosis and in the environment.14,31,36
The C. gattii strain isolated from Grevillea robusta was type VGI. This finding agrees with that reported by Refojo et al. and Davel et al., who isolated C. gattii, though from other tree species in Buenos Aires city.9,36 In other South American countries, such as Brazil and Colombia, the most common genotypes were VGII and VGIII.14,31,36,39,43 This would demonstrate the geographic variation in the distribution of molecular types of C. gattii.
The study of the genetic relationship between environmental strains of Cryptococcus and those isolated from clinical specimens would be interesting to confirm that trees are a reservoir or source of infection.
This is the first study about ecological niches of Cryptococcus carried out in the northeast of Argentina. Further researches are necessary to find out which are the circulating genotypes in South America.
Conflict of interestThe authors report no conflict of interest. The authors alone are responsible for the content and writing of the paper.
Financial support for this work was provided by Fundación Alberto J Roemmers.