Articles in press
"Articles in press" son artículos aceptados y revisados pero que aún no han sido asignados a un volumen/número. Pueden ser citados usando el DOI - Saber más
- •
Pseudomycetoma is a rare chronic granulomatous reaction that surrounds fungal hyphae.
- •
Mycobacterium genavense does not grow on conventional solid and liquid media.
- •
Severely immunosuppressed patients may present co-infections.
- •
En agua de mar de la costa marplatense se confirmó la presencia de Acanthamoeba spp.
- •
Acanthamoeba spp. sobrevive a condiciones ambientales diversas por su adaptabilidad.
- •
Se reportó por primera vez Acanthamoeba spp. en agua de mar en Argentina.
- •
Candida can colonize and invade human surfaces, causing severe infections.
- •
Incidence of candidemia in our study was higher than Latin American.
- •
Non-albicans Candida species were the most frequent, including C. parapsilosis.
- •
Septic shock was the only one with a risk factor for mortality.
- •
Implementing infection control strategies to prevent candidemia and adverse outcomes.
- •
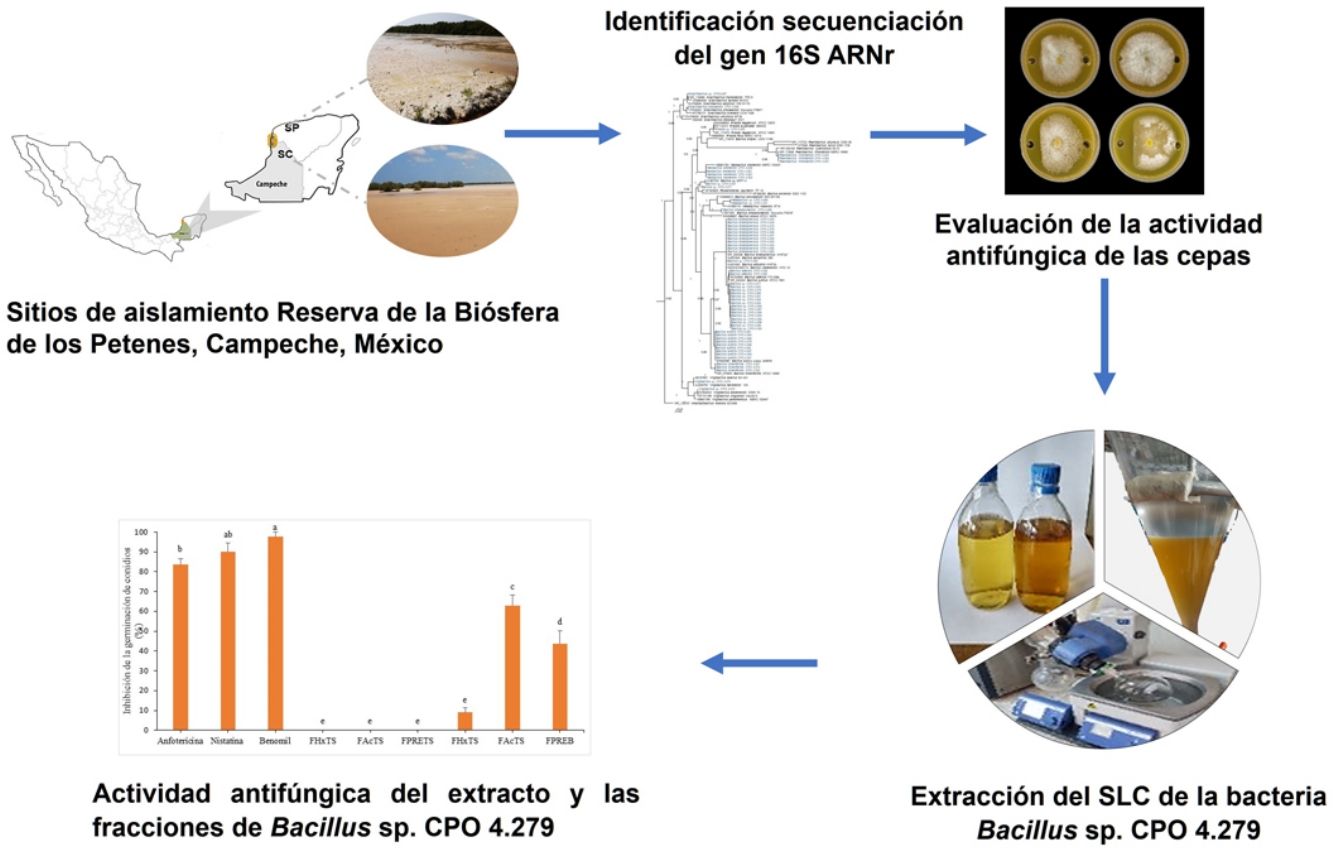
Los suelos salinos blanquizales poseen bacterias con propiedades antifúngicas.
- •
Bacillus sp. CPO 4.279 presentó la actividad antifúngica más alta.
- •
El extracto EcB9 mostró actividad significativa contra Colletotrichum gloeosporioides.
- •
La fracción de mediana polaridad FAcB9 posee metabolitos secundarios antifúngicos.
- •
Filamentous fungi in hospital indoor air are risk for severely neutropenic patients.
- •
Monitoring microbiology quality air to prevent invasive fungal diseases.
- •
Importance of mycology laboratory in air monitoring bone marrow transplant units.
- •
Fermented low-alcoholic beverages have special flavor and health benefits.
- •
Health benefits of alcoholic fermented beverages are often attributed to peptides.
- •
Lactic acid bacteria exhibit a proteolytic system that release bioactive peptides.
- •
Bioactive peptides are also release by yeast metabolism and its autolysis.
- •
Fructanos de agave en una concentración del 0,1%, presentan actividad prebiótica
- •
El 0,1% de fructanos de agave estimula la formación de biopelículas en probióticos
- •
Fructanos de agave contribuye a generar estructuras que favorecen la colonización de probióticos
- •
First report of the sexual phase of Sclerotinia sclerotiorum in Patagonia - Argentina.
- •
High incidence of ascosporic infections.
- •
The inhibition of antibiotics is detected with a simple bioassay in a short time.
- •
Cephalosporins, quinolones and tetracyclines inhibits the growth of K. marxianus.
- •
Similar bioassays could analyze the antibiotic susceptibility of pathogenic yeasts.
- •
Kids are not vaccinated against brucellosis.
- •
Strain B. melitensis Rev 1 ΔeryCD was not isolated from vaccinated kids.
- •
Rev 1 vaccine was only isolated from 1/15 vaccinated kids.
- •
Vaccination in males does not cause lesions in the reproductive organs.





