The increase in the incidence of pancreatic and biliary cancers has attracted the search for methods of early detection of diseases and biomarkers. The authors propose to analyze new findings on the association between microbiota and Pancreatic Ductal Adenocarcinoma (PDAC) or Cholangiocarcinoma (CCA).
MethodsThis systematic review was carried out according to the items of Preferred Reports for Systematic Reviews and Protocol Meta-Analysis (PRISMA-P). This study was registered by the Prospective Register of Systematic Reviews (PROSPERO), identification code CRD42020192748 before the review was carried out. Articles were selected from the PUBMED, EMBASE, and Cochrane databases.
ResultsMost studies (86.67%) used 16s rRNA as a sequencing method. The main comorbidities found were diabetes mellitus, systemic arterial hypertension, and dyslipidemia. Many studies were limited by the small number of participants, but the biases were mostly low. There was very little concordance about the composition of the microbiome of different sites, for both case and control groups when compared to other studies' results. Bile sample analysis was the one with a greater agreement between studies, as three out of four studies found Escherichia in cases of CCA.
ConclusionThere was great disagreement in the characterization of both the microbiota of cases and control groups. Studies are still scarce, making it difficult to adequately assess the data in this regard. It was not possible to specify any marker or to associate any genus of microbiota bacteria with PDAC or CCA.
The incidence and prevalence of cancer have increased over time.1 Pancreatic cancer, mainly Pancreatic Ductal Adenocarcinoma (PDAC), is the fourth type of cancer with the highest overall mortality in the United States2 and, in 2018, was responsible for about 4.6% of cancer deaths worldwide,3 with 4.8% in Brazil.4 Bile duct cancer has a low incidence in the Western world (between 0.35 and 2 per 100,000 per year). Due to the initial silent progression and the difficulty of early detection, both diseases have a poor prognosis, with five years survival of less than 20%.5,6 Screening is restricted to individuals at high risk of developing the diseases,7 which are often discovered in advanced, metastatic stages, as there are no large-scale or non-invasive screening tests available.
The etiology of these cancers is still not well defined. Among the most mentioned causes, there are chronic inflammation, genomic factors, biliary cysts, viral infections and, recently, alterations in the microbiota, called dysbiosis.8
Despite the difficulty in determining what constitutes a microbiota in eubiosis or dysbiosis, some associations can be established. Del Castillo et al.,9 found a reduction in the presence of Lactobacillus and an increase in Porphyromonas in cases of PDAC compared to the control group. Wei M-Y et al. found an association of Porphyromonas gingivalis, which causes chronic periodontitis, with the development of PDAC, through the expression of peptidyl-arginine-deiminase, which promotes mutations in K-ras and p53. The relationship between the presence of Helicobacter pylori, a bacterium associated with malignant transformation in the stomach, and the development of PDAC is also being studied. However, its participation as a risk factor for the disease is not yet proven, in addition to being very controversial.10-13
Regarding CCA, despite the small number of studies on the subject, some demonstrate that the presence of H. pylori, H. bilis and H. hepaticus in the intestine leads to an increase in the Nuclear Factor Kappa B (NFKB) and nuclear signaling pathway production of Vascular Endothelial Growth Factor (VEGF). Thus, there would be an increased risk for the development of neoplasms, with angiogenesis promotion in the tumor site.14
When considering the relationship between cancer and changes in the microbiota, there is a possibility of tracking the disease. Farrel et al.,10 after finding a reduction in bacteria from the oral microbiota, Neisseria elongata and Streptococcus mitis, in patients with PDAC, suggested that studying the oral microbiota could be used as screening for pancreatic neoplasms. Thus, there are a variety of sites to be potentially explored for a better understanding of this disease, not only the pancreas itself.
The mechanism of colonization of the bile ducts and pancreas is not yet defined and is still a topic under discussion.15,16 Given the high morbidity and mortality of PDAC and CCA and the difficulties of early detection, it is essential to develop methods for screening the disease and discovering biomarkers.
ObjectiveThe aim of this systematic review is to evaluate new findings and reports on the composition of the gastrointestinal tract microbiota in cases of pancreatic and biliary cancer.
MethodsThis systematic review was carried out according to the items of Preferred Reports for Systematic Reviews and Protocol Meta-Analysis (PRISMA-P).17 This study was registered by the Prospective Register of Systematic Reviews (PROSPERO, identification code CRD42020192748) before the review was carried out.
The preparation of the research question was based on the PICO strategy,18 considering diseases of the pancreas and biliary tree (Patient or Problem); microbiota impact (Interest); healthy people or people with benign diseases (Control or Comparison); all outcomes available in the literature were c onsidered in the analysis (Outcome).
Eligibility criteriaTypes of studiesArticles were selected from their titles and abstracts according to their data relevance and regardless of their publication status. Articles with full text inaccessible to authors were not considered.
The following study designs were considered: randomized controlled clinical trials, non-randomized controlled clinical trials, prospective and retrospective cohorts, case-control and cross-sectional. Reports and case series, reviews, letters to the editors, research protocols and congress proceedings were not considered.
Types of participantsStudy participants were adults with pancreatic or bile duct cancer and control subjects who underwent gastrointestinal microbiota evaluation.
Methods of sample collectionThe sample collection strategies evaluated in the study are stool analysis, ERCP, oral swab, surgery for biliopancreatic disease and upper digestive endoscopy.
Types of variables/parameters analyzedData relating to the authors, date, and location (country) of publication, type of study, analysis methods, analyzed site, associated factors and microbiota alteration were collected and arranged in tables.
Exclusion criteriaStudies were excluded if: samples were collected from children, adolescents, patients with autoimmune disease, cadavers, or rodents; the article is incomplete or unpublished; tumors that are not primarily of the pancreas or biliary tree; are in other languages except English and Portuguese.
Literature reviewThe survey was conducted on August 11, 2021, without language or date restrictions, in the following databases: Medline (via PubMed) ‒ www.pubmed.com; EMBASE ‒ www.embase.com and Cochrane ‒ www.cochranelibrary.com.
Using the PubMed search tool, the authors selected MeSH terms from the most relevant publications to perform a new search, in order to obtain articles that could be included in this systematic review.
In addition, a manual search of theses, meetings, references, study records, and contact with experts in the field was carried out.
Search strategyThe keywords were used equally in all databases, respecting their heterogeneities (for example, Emtree terms and MeSH terms were mapped in Embase and Medline, respectively).
The keywords were: “pancreatic neoplasms”, “microbiota”, “early detection of cancer”; “dysbiosis”, “tumor microenvironment” and “biliary tract neoplasms”.
The search strategy was: ((Microbiota) or (Dysbiosis)) and ((Pancreatic Neoplasms) or (Biliary Tract Neoplasms) or (Early Detection of Cancer) or (Tumor Microenvironment)).
Data extractionData from each study were independently extracted by four authors (V.C.M., E.M.C.D., M.O.S and F.S.N.). Disagreements were resolved by consensus. If no consensus was reached, a fifth author (A.M.) would be consulted. Data extraction was carried out using the Rayyan tool ‒ https://rayyan.qcri.org/.19
All studies were analyzed according to their titles and abstracts, according to inclusion and exclusion criteria. If the eligibility criteria were met, the full text would be extracted. All evaluated full-text studies were described in the “Results” section.
Missing data were clarified by contacting the authors directly.
Data validationThe four authors performed data validation through the discussion of selected works. If no consensus was reached, a fifth author would be consulted.
The risks of bias for the studies were assessed using the Study Quality Assessment Tools | National Heart, Lung, and Blood Institute (NHLBI).20 Intervention-type studies were analyzed using the guidelines of the Cochrane Back Review Group (CBRG).21
All selected studies were considered.
Authors' responsibilities/contributionsV.C.M., M.O.S. and F.S.N.: Conception, methodology, formal analysis, investigation, writing, drafted the work; J.V.T. and L.Z.P.: Validation, review; AM: Conception, methodology, investigation, supervision, project administration.
All authors have approved the submitted version and have agreed to both be personally accountable for the author's own contributions and to ensure that questions related to the accuracy or integrity of any part of the work, even ones in which the author was not personally involved, are appropriately investigated, resolved, and the resolution documented in the literature.
ResultsSearch flow1577 results were found for the keywords used. A total of 308 duplicates were removed, leaving 160 potentially eligible studies after abstract analysis. Of those selected, 122 were not included for not meeting the inclusion criteria, 5 studies were excluded for using rodents, 1 study for inclusion of children, 1 for using a cadaver, 1 for treating an autoimmune disease associated with cancer, 1 for study in a liver fluke endemic area, 1 for blood serology analysis, 2 for use of biobank material and 11 not related to the review question. In the end, 15 studies were selected for qualitative analysis (Fig. 1).
Limitations and methodologies of studiesAmong the limitations reported in the studies, the small size of the population sample, the single collection of the material to be analyzed, and collection after cancer diagnosis were the most frequent (Table 1).
Reported studies limitations.
Fan, X et al.22 used saliva samples collected prior to suspicion and diagnosis of PDAC. Thus, results were obtained without the potential influence of the disease on the microbiome composition. However, post-diagnosis samples were not collected to identify whether there are changes in the microbiota caused by this condition.
Quality of evidenceThe articles selected for this review are studies that collect material (saliva, feces, bile, tissue, and duodenal and bile duct fluid) from patients and controls, and analyze the microbiota present in each sample. After analyzing the studies, the selection, detection, reporting, information, and loss biases were observed to define the quality of the evidence found, with the classification displayed in Figs. 2 and 3.
The studies included in this review show mostly low selection bias, with 11 being low and 4 uncertain. Among the uncertain case-control studies, Torres, P.J. et al.,23 determines the ethnicity of its participants but do not restrict the location of the population in question. Ren, Z. et al.24 describes the country of origin analyzed, but does not describe ethnic and socioeconomic characteristics.
The cross-sectional study by Chen, B. et al.25 and the cohort of Riquelme E., et al.26 had low selection bias. The first one selected and recruited patients from the same population, that underwent the ERCP procedure between 2016 and 2017 at the Shanghai General Hospital, and both specified and uniformly applied inclusion and exclusion criteria in the selection.
The cross-sectional study by Serra, N. et al.27 and the cohort of Di Carlo, P. et al.28 had an uncertain selection bias, as it did not report the exclusion criteria, and it was not possible to determine whether these were applied uniformly to all participants.
Furthermore, the studies present an uncertain detection bias, as they do not report whether or not there was a blinding in the performance of the analyses. This cannot be applied to Vogtmann, E. et al.29 and Q.-X. Mei et al.,30 showed low detection bias. Vogtmann, E. et al. performed a blinded analysis of each sample, whether case or control, specifying this in the methodology. Q.-X. Mei et al., however, did not clarify regarding the blinding in the analysis of the microbiome of each study participant, only the blinding of pathologists in the histological analysis of duodenal tissue samples.
As the composition of the microbiome is a non-self-reported characteristic, without the possibility of being influenced by factors such as patient memory, omission, or addition of data, all articles analyzed in this review had low reporting bias.
The low information bias, shared by all of the articles considered, is due to the clear criteria used to separate the groups. The division was made based on the histopathological diagnosis of the presence or absence of cancer, reducing the wrong allocation.
The low loss bias remains on the fact that most studies performed a single analysis of the microbiome without the need for follow-up, both with patients and controls. However, two cohorts showed unclear loss bias (Di Carlo, P. et al.28 and Riquelme E. et al.26), due to the fact that both analyze the survival of selected patients. Thus, for this review, only the data collected at admission were used.
Characteristics of the studiesThe demographic characteristics collected were displayed in Table 2; the main changes, results and conclusions are provided in Tables 3-5.
Demographic characteristics of studies.
| Author, publication date and country | Number of patients | Mean age ‒ years (SD) or Range of age (n) | Sex (%) | Associated factors (n or %) |
|---|---|---|---|---|
| Chen B (2019), China | CCA: 8 | CCA: 72.13 (range: 60‒95) | CCA: 3 M (37.5); 5 F (62.5) | DM: CBD stones: 10, RC: 4 |
| CBD stones: 44 | CBD stones: 66.98 (range: 44‒90) | CBD stones: 18 M (40.9); 26 F (59.1) | Dyslipidemia: CCA: 1, CBD stones: 15, RC: 4 | |
| RC: 16 | RC: 73.94 (range: 49‒92) | RC: 9 M (56.2); 7 F (43.8) | Hypertension: CCA: 5, CBD stones: 17, RC: 6 | |
| Total: 68 | Elevated ALT or AST: CCA: 7, CBD stones: 25, RC: 7 | |||
| Elevated Tbil and/or Dbil: CCA: 6, CBD stones: 19, RC: 7 | ||||
| Elevated Scr: CCA: 2, CBD stones: 6, RC: 2 | ||||
| Elevated WBC or NE: CBD stones: 7 | ||||
| Cholecystolithiasis: CCA: 1, CBD stones: 23, RC: 7 | ||||
| Vogtmann E (2020), USA | PDAC: 273 | PDAC: < 50 (26), 50‒59 (61), 60‒69 (87), 70‒79 (77), ≥ 80 (22) | PDAC: 165 M (60.4), 108 F (39.6) | ‒ |
| Control group: 285 | Control group: 131 M (46); 154 F (54) | |||
| Control group: < 50 (20), 50‒59 (78), 60‒69 (94), 70‒79 (59), ≥ 80 (34) | ||||
| Total: 558 | ||||
| Half E (2019), Israel | Case group: PDAC: 30, PCL: 6 | PDAC: 69.9 (6.2) | PDAC: 16 M (53.5), 14 F (46.7) | DM: PDAC: 53%, PCL: 20%, NAFLD: 13% |
| Control group: NAFLD: 16 | PCL: 66 (15.3) | PCL: 5 M (83.3), 1 F (16.7) | Hypertension: PDAC: 43%, PCL: 25%, NAFLD: 50% | |
| Healthy control: 13 | NAFLD: 51 (10.8) | NAFLD: 12 M (75), 4 F (25) | ||
| Total: 65 | Healthy control: 59 (8.7) | Healthy control: 6 M (46.6), 7 F (53.4) | Dyslipidemia: PDAC: 40%, PCL: 29%, NAFLD: 88%, Healthy control: 23% | |
| Bile-duct obstruction: PDAC: 36% | ||||
| Gall-bladder abnormalities: PDAC: 46%, NAFLD: 6%, Healthy control: 23% | ||||
| Serra N (2018), Canada | CCA: 20, GBC: 2, PDAC: 31, Total: 53 | 73.4 (10.5) | 0 M (0), 53 F (100) | Intra-abdominal infection: 53 |
| Olson SH (2017), USA | PDAC: 40a | PDAC: < 60 (10), 60‒69 (12), ≥ 70 (12) | PDAC: 18 M (53), 16 F (47) | DM: PDAC: 9, IPMN: 4, Healthy control group: 7 |
| IPMN: 39 | ||||
| Healthy control group: 58 | IPMN: < 60 (8), 60‒69 (8), ≥ 70 (23) | IPMN: 22 M (40), 17 F (60) | Álcool: PDAC: 25, IPMN: 36, Healthy control group: 53 | |
| Total: 137 | Healthy control group: < 60 (20), 60‒69 (7), ≥ 70 (11) | Healthy control group: 23 M (56), 35 F (44) | Gum disease ever: PDAC: 14, IPMN: 15, Healthy control group: 19 | |
| Di Carlo P (2019), Italy | PDAC: 72 | PDAC: 75.6 (10.4) | PDAC: 41 M (56.9), 31 F (43.1) | ‒ |
| CCA: 39 | ||||
| Total: 111 | CCA: 71.5 (8.8) | CCA: 19 M (48.7), 20 F (51.3) | ||
| Mei Q-X (2018), China | PDAC: 14 | PDAC: 56.8 (5.1) | PDAC: 9 M (64.3), 5 F (35.7) | ‒ |
| Control group: 14 | ||||
| Control group: 55.4 (6.2) | Control group: 9 M (64.3), 5 F (35.7) | |||
| Total: 28 | ||||
| Riquelme E (2019), USA | PDAC Discovery cohort (DC): LTS: 21, STS: 22 | DC: LTS: 62.71 (range: 44‒73), STS: 62.05 (range: 46‒74) | DC: LTS: 10 M (47.62), 11 F (52.38); STS: 13 M (59.1); 9 F (41.9) | ‒ |
| Validation cohort (VC): LTS: 15, STS: 10 | ||||
| Torres PJ (2015), USA | PDAC: 8 | PDAC: 71.1 | PDAC: 6 M (75), 2 F (25) | Other diseases group: Non-pancreatic cancer, Pancreatic diseases (not cancer) |
| Others diseases: 78 | Others diseases: no descript | |||
| Others diseases: 38 M (48.72), 40 F (51.28) | ||||
| Healthy control group: 22, | Healthy control group: no descript | |||
| Healthy control group: 12 M (54.55), 10 F (45.45) | ||||
| Total: 108 | ||||
| Ren Z (2017), China | PDAC: 85 | PDAC: 56 (range: 33‒78) | PDAC: 47 M (55.3), 38 F (44.7) | ‒ |
| Healthy control group: 57 | Healthy control group: 52 (range: 43‒67) | Healthy control group: 36 M (63.2), 21 F (36.8) | ||
| Total: 142 | ||||
| Fan X (2018), USA | Group 1: PDAC: 170, Control group: 170 | Group 1: PDAC: 73.7 (5.7), Control group: 73.7 (5.7) | Group 1: PDAC: 90 M (52.9), 80 F (47.1), Control group: 90 M (52.9), 80 F (47.1) | ‒ |
| Group 2: PDAC: 191, Control group: 201 | Group 2: PDAC: 63.8 (5.2), Control group: 633.8 (5.4) | Group 2: PDAC: 116 M (60.7), 75 F (39.3), Control group: 122 M (60.7), 79 F (39.3) | ||
| Total: 732 | ||||
| Kohi S (2021), USA | Control: 134 | Control: 63.6 (41.6–79.5) | Control: 30 M (47.6%), 33 F (52.4%) | ‒ |
| PDAC: 74 | ||||
| Cyst: 98 | PDAC: 42.2–85.5 | PDAC: M 64%, F 36% | ||
| Total: 308 | Cyst: 65.8 (42.9–87.8) | Cyst: 36 M (50%), 36 F (50%) | ||
| Saab M (2021), France | CCA: 28 | CCA: 64 (12) | CCA: 19 M (68), 9 F (32) | DM: CCA: 6, Biliary duct lithiasis: 9 |
| Biliary duct lithiasis: 47 | Biliary duct lithiasis: 57 (17) | Biliary duct lithiasis: 23 M (49), 24 F (51) | Pancreatitis: CCA: 0, Biliary duct lithiasis: 1 | |
| Total: 75 | Inflammatory bowel disease: CCA: 2, Biliary duct ithiasis: 0 | |||
| Primary sclerosing cholangitis: CCA: 1, Biliary duct lithiasis: 0 | ||||
| Sun H (2020), China | PDAC: 10 | PDAC: 57.4 (7.8) | PDAC: 6 M (60), 4 F (40) | Alcool: PDAC: 2, BPD: 3, Healthy control group: 1 |
| BPD: 17 | BPD: 42.8 (16) | |||
| Healthy control group: 10 | Healthy control group: 31.1 (2.7) | BPD: 10 M (58.82), 7 F (41.17) | Smoking: PDAC: 2, BPD: 3, Healthy control group: 2 | |
| Total: 37 | Healthy control group: 6 M (60), 4 F (40) | BPD: Pancreatitis, chronic pancreatitis, benign pancreatic tumors | ||
| Wei AL (2020), China | PDAC: 41 | PDAC: 61.17 (1.79) | PDAC: 24 M (59), 17 F (41) | DM: PDAC: 5, Healthy control group: 11 |
| Healthy control group: 69 | Healthy control group: 64.64 (1.04) | Healthy control group: 50 M (72), 19 F (28) | Hypertension: PDAC: 4, Healthy control group: 20 | |
| Total: 110 | Alcool: PDAC: 16, Healthy control group: 30 | |||
| Smoking: PDAC: 17, Healthy control group: 37 |
M, Male; F, Female; CBD stones, Common bile duct stones; RC, Recurrent Choledocholithiasis; PC, Pancreatic Cancer; PCL, Precancerous Lesions; NAFLD, Non-alcoholic Fat Liver Disease; CCA, Cholangiocarcinoma; GBC, Gallbladder Carcinoma; PDAC, Pancreatic Ductal Adenocarcinoma. IPMN, Intraductal Papillary Mucinous Neoplasms; DM, Diabetes Mellitus; AST, Aspartate Aminotransferase; ALT, Alanine Aminotransferase; Tbil, Total Bilirubin; Dbil, Direct Bilirubin; Scr, Serum Creatinine; WBC, White Blood Cell; NE, Neutrophilic Granulocyte.
Experimental studies main results.
| Author, publication date and country | Study type | Analysis Method | Type of disease | Type of sample | Microbiota (caso/controle) | |
|---|---|---|---|---|---|---|
| Vogtmann E (2020), USA | Case-control | 16s rRNA | Case: PDAC | Saliva | Phylum: Bacteroidetes, Firmicutes, Proteobacteria | Phylum: Proteobacteria |
| Control: Submucosal lesions in the esophagus or stomach, Cholelithiasis or choledocholithiasis without cholangitis. | Family: Pasteurellaceae | |||||
| Family: Bacteroidaceae, Staphylococcaceae, Enterobacteriaceae, Lachnospiraceae | Genus: Haemophilus | |||||
| Genus: Lachnospiraceae G7 | ||||||
| Half E (2019), Israel | Case-control | 16s rRNA | PDAC | Stool | PDAC (compared with healthy individuals from the Israeli cohort): Phylum: Firmicutes, Verrucomicrobia, Bacterioidetes, Family: Veillonellaceae, Akkermansiaceae, Odoribacteraceae, Genus: Megasphaera, Akkermansia, Odoribacter | Healthy (compared to individuals with PDAC in the Israeli cohort): Phylum: Firmicutes, Tenericutes, Family: Clostridiacea, Lachnospiraceae, Erysipelotrichaeceae, Ruminococcaceae, Genus: Clostridium, Anaerostipes, Faecalibacterium, Subdoligranulum |
| PDAC (compared to individuals with PDAC in the Chinese cohort): Phylum: Firmicutes, Family: Veillonellaceae | Healthy (compared to individuals with PDAC in the Chinese cohort): Phylum: Firmicutes, Family: Erysipelotrichaeceae, Clostridiaceae, Genus: Anaerostipes | |||||
| Olson SH (2017), USA | Case-control | 16s rRNA | Case: PDAC | Saliva | PDAC (compared to healthy control or IPMN): Phylum: Firmicutes, Family: Streptococcaceae, Genus: Streptococcus | Healthy control (compared with PDAC): Phylum: Proteobacteria, Family: Pasteurellaceae, Neisseriaceae, Genus: Haemophilus, Neisseria |
| Control: IPMN | ||||||
| Mei Q-X (2018), China | Case-control | 16s rRNA | PDAC | Duodenum | PDAC (compared to healthy control): Phylum: Proteobacteria, Firmicutes, Deinococcus-Thermus; Family: Moraxellaceae, Comamonadaceae, Yersiniaceae, Comamonadaceae, Sphingomonadaceae, Bacillaceae, Deinococcaceae, Oxalobacteraceae; Genus: Acinetobacter, Aquabacterium, Rahnella, Delftia, Sphingobium, Massilia, Oceanobacillus, Deinococcus | Control (compared to PDAC): Phylum: Proteobacteria, bacterioidetes, firmicutes; Family: Enterobacteriaceae, Pseudomonadaceae, Incertae sedis, Phophyromonadaceae, Paenibacillaceae; Genus: Escherichia, Shigella, Pseudomonas, Enhydrobacter, Porphyromonas, Paenibacillus |
| Most Abundant PDAC: Phylum: Firmicutes, Proteobacteria, Bacteroidetes; Family: Bacillaceae, Pseudomonadaceae, Streptococcaceae; Genus: Bacillus, Pseudomonas, Lactococcus | More abundant healthy: Phylum: Firmicutes, Proteobacteria, Bacteroidetes; Family: Bacillaceae, Pseudomonadaceae, Streptococcaceae; Genus: Bacillus, Pseudomonas, Lactococcus | |||||
| Torres PJ (2015), USA | Case-control | 16s rRNA | PDAC | Saliva | Increase in PDAC (compared to other diseases and healthy controls): Phylum: Fusobacteria, Family: Leptotrichiaceae, Genus: Leptotrichia | No descript |
| Decrease in PDAC (compared to other diseases and healthy controls): Phylum: Bacteroidetes, Proteobacteria, Family: Porphyromonadaceae, Neisseriaceae; Genus: Porphyromonas, Neisseria | ||||||
| Ren Z (2017), China | Case-control | 16s rRNA | PDAC | Stool | PDAC (compared with healthy control group): Phylum: Bacteroidetes; Family: Prevotellaceae, Veillonellaceae, Enterobacteriaceae; Genus: Prevotella, Hallella, Veillonella, Selenomonas, Klebsiella, Enterobacter, Cronobacter | Healthy control group (compared with PDAC): Phylum: Firmicutes, Proteobacteria, Actinobacteria; Family: Ruminococcaceae, Lachnospiraceae, Clostridiaceae, Bifidobacteriaceae; Genus: Gemmiger, Flavonifractor, Coprococcus, Blautia, Anaerostipes, Clostridium IV, Butyricicoccus, Dorea, Bifidobacterium |
| Most Abundant PDAC: Phylum: Bacteroidetes, Firmicutes e Proteobacteria | More abundant healthy control group: Phylum: Bacteroidetes, Firmicutes e Proteobacteria | |||||
| Fan X (2018), USA | Case-control | 16s rRNA | PDAC | Saliva | Associated with high risk of pancreatic cancer: Phylum: Bacterioidetes1, Proteobacteria2; Family: Porphyromonadaceae1, Pasteurellaceae2; Genus: Porphyromonas1, Aggregatibacter2; aEspécie: Porphyromonas gingivalis1, Aggregatibacter actinomycetemcomitans2 | Associated with low risk of pancreatic cancer: Phylum: Fusobacteria; Family: Leptotrichiaceae; Genus: Leptotrichia |
| Kohi S (2021), USA | Case-control | 16s rRNA | PDAC | Duodenal fluid | Most Abundant PDAC: Phylum: Proteobacteria | Phylum: Firmicutes, Bacteroidetes, Proteobacteria: Class: Bacilli, Bacteroidia, Negativicutes, Gammaproteobacteria; Order: Lactobacillales, Bacteroidales, Selenomonadales; Familia: Streptococcaceae, Veillonellaceae, Prevotellaceae; Genus: Streptococcous, Veillonella, Prevotella 7 |
| Pancreatic cyst | ||||||
| PDAC (compared with healthy control group): Phylum: Actinobacteria, Fusobacteria, Firmicutes; Family: Bifidobacteriaceae, Lactobacillaceae, Enterococcaceae; Genus: Bifidobacterium, Fusobacterium, Enterococcus | ||||||
| PDAC (compared with pancreatic cyst): Phylum: Proteobacteria, Bifidobacterium, Enterococcus; Genus: Escherichia-Shigella, Clostridium sensu strictu, Enterococcus, Bifidobacterium | Cyst: Family: Porphyromonadaceae, Corynebacteriaceae, Leptotrichiaceae; Genus: Streptococcus, Veillonella, Prevotella | |||||
| STS: Class: Fusobacteria, Actinobacteria, Betaproteobacteria; Genus: Fusobacterium, Rothia, Neisseria | There was no difference between cyst and control | |||||
| Saab M (2021), France | Case-control | 16s rRNA | CCA | Bile | CCA (compared with biliary lithiasis): Phylum: Bacteroidetes; Family: Bacteroidaceae; Genuss: Streptococcus, Bacteroides e Pyramidobacter | Biliary lithiasis (compared with CCA): Phylum: Firmicutes; Family: Clostridiaceae, Enterobacteriaceae, Fusobacteriaceae, Enterococcaceae; Genus: Clostriduim, Klebsiella, Fusobacterium e Enterococcus |
| Biliary lithiasis | ||||||
| Sun H (2020), China | Case-control | 16s rRNA | PDAC | Saliva | PDAC e BPD (compared with Healthy control group): Phylum: Spirochaetes; Family: no important variation | Healthy control group (compared with PDAC e BPD): Phylum: Proteobacteria; Family: Neisseriaceae |
| BPD (benign pancreatic disease) | ||||||
| Most Abundant in PDAC cases: Phylum: Bacteroidetes; Family: Prevotellaceae, Genus: Neisseria, Veillonella | Most Abundant in Healthy control group: Phylum: Proteobacteria; Family: Neisseriaceae; Genus: Neisseria | |||||
| Wei AL (2020), China | Case-control | 16s rRNA | PDAC | Saliva | Increased in PDAC cases (compared with Healthy control group): Phylum: Firmicutes, Fusobacteria, Actinobacteria, Proteobacteria; Family: Streptococcaceae, Lactobacillaceae, Leptotrichiaceae, Actinomycetaceae, Micrococcaceae, Enterobacteriaceae; Genus: Streptococcus, Lactobacillus, Leptotrichina, Actinomyces, Rothia, Escherichia | Reduced in PDAC cases (compared with Healthy control group): Phylum: Proteobacteria, Firmicutes, Bacteroidetes; Family: Neisseriaceae, Veillonellaceae, Porphyromonadaceae, Flavobacteriaceae, Prevotellaceae; Genus: Neisseria, Selenomonas, Porphyromnas, Tannerella, Capnocytophaga, Alloprevotella |
| Associated with high risk of pancreatic cancer: Phylum: Firmicutes, Fusobacteria; Family: Streptococcaceae, Leptotrichiaceae; Genus: Streptococcus, Leptotrichia | Associated with low risk of pancreatic cancer: Phylum: Proteobacteria, Bacteroidetes; Family: Neisseriaceae, Prevotellaceae; Genus: Neisseria, Veillonella | |||||
PDAC, Pancreatic Ductal Adenocarcinoma; IPMN, Intraductal Papillary Mucinous Neoplasms. STS, Short-Term Survival.
Observational studies main results.
| Author, publication date and country | Type of study | Analysis method | Type of disease found | Type of sample | Microbiota found (case/control) | |
|---|---|---|---|---|---|---|
| Chen B (2019), China | Cross-sectional | 16s rRNA | Case: CCA | Bile | CCA: Phylum: Gemmatimonadetes, Nitrospirae, Chloroflexi, Latescibacteria, and Planctomycetes, Proteobacteria1, Firmicutes2, Actinobacteria3; Family: Enterobacteriaceae1, Staphylococcaceae2.1, Ruminococcaceae2.2, Microbacteriaceae3.1, Corynebacteriaceae3.2; Genus: Escherichia/Shigella1, Klebsiella1, Staphylococcus2.1, Unclassified_Enterobacteriaceae, and Faecalibacterium2.2, Okibacterium3.1, and Corynebacterium3.2 | CBD stones: Phylum: Proteobacteria1, Firmicutes2; Family: Enterobacteriaceae1.1, Halomonadaceae1.2, Streptococcaceae2; Genus: Escherichia/Shigella1.1, Klebsiella1.1, Enterococcus1.1, Halomonas1.2, Streptococcus2 |
| Control: CBD stones | ||||||
| Serra N (2018), Canada | Cross-sectional | BD Phoenix system | CCA | Bile | PDAC: Phylum: Proteobacteria; Family: Enterobacteriaceae; Genus: Klebsiella | |
| GBC | ||||||
| PDAC | ||||||
| CCA: Phylum: Proteobacteria; Family: Pseudomonadaceae e Enterobacteriaceae; Genus: Pseudomonas e Escherichia | ||||||
| GBC: Phylum: Proteobacteria; Family: Pseudomonadaceae; Genus: Pseudomonas | ||||||
| Di Carlo P (2019), Italy | Cohort | BD Phoenix system or Vitek-2 System | CCA | Bile | PDAC: Phylum: Proteobacteria1; Family: Enterobacteriaceae1.1, Pseudomonadaceae1.2; Genus: Escherichia1.1, Klebsiella1.1, Pseudômonas 1.2. | |
| PDAC | ||||||
| CCA: Phylum: Proteobacteria1; Family: Enterobacteriaceae1.1, Pseudomonadaceae1.2; Genus: Escherichia1.1, Pseudômonas 1.2 | ||||||
| Riquelme E (2019), USA | Cohort | 16s rRNA | PDAC | Stool | LTS: Phylum: Proteobacteria1, Actinobacteria2; Class: Xanthomonadaceae1, spretomycetales2.1, Pseudonocardiaceae2.2, Baccilaceae3; Genus: Pseudoxanthomonas1, Streptomyces2.1, Saccharopolyspora2.2; bSpecies: Bacillus clausii | |
| STS: Phylum: Firmicutes1, Bacterioidetes2; aClass: Clostridia1, Bacteroide2 | ||||||
CCA, Cholangiocarcinoma; CBD stones, Common bile Duct Stones; GBC, Gallbladder Carcinoma; PDAC, Pancreatic Ductal Adenocarcinoma; LTS, Long Term Survival; STS, Short Term Survival.
Objectives and conclusions of studies.
CCA, Cholangiocarcinoma; PC, Pancreatic Cancer; IPMN, Intraductal Papillary Mucinous Neoplasms; PBD, Pancreas and Biliary Tract Disease; PDAC, Pancreatic Ductal Adenocarcinoma; LTS, Long Term Survival; HTS, High-Throughput Sequencing; ERCP, Endoscopic Retrograde Cholangiopancreatography.
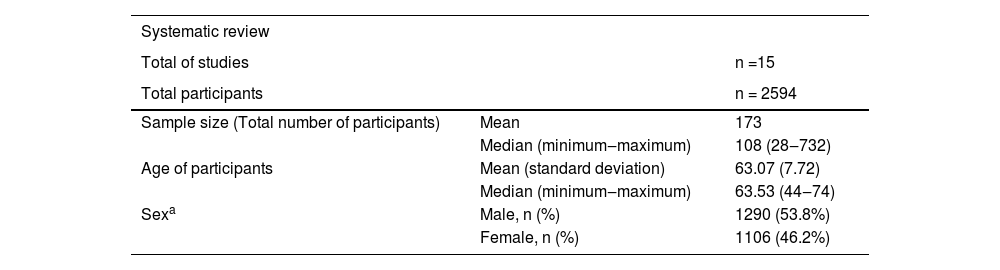
15 scientific papers were included, with a total of 2594 participants. The minimum number of participants in a study was 28 and the maximum was 732, 50% of the studies had at least 108 participants. The mean age of participants was 63.07 years (standard deviation 7.72). Most participants were male (53.8%). The results are shown in Table 6.
Description of studies included in the systematic review.
| Systematic review | ||
|---|---|---|
| Total of studies | n =15 | |
| Total participants | n = 2594 | |
| Sample size (Total number of participants) | Mean | 173 |
| Median (minimum‒maximum) | 108 (28‒732) | |
| Age of participants | Mean (standard deviation) | 63.07 (7.72) |
| Median (minimum‒maximum) | 63.53 (44‒74) | |
| Sexa | Male, n (%) | 1290 (53.8%) |
| Female, n (%) | 1106 (46.2%) |
The comorbidities listed in the articles mainly include diabetes mellitus, systemic arterial hypertension, and dyslipidemia. Riquelme E. et al.26 and Half, E. et al.31 include obstruction of the biliary tract (caused by the presence of tumor, calculi, thickening of walls, or unknown reasons) due to increased serum levels of canalicular enzymes. Other associated factors are alcohol consumption and smoking, increased serum creatinine and white blood cells, cholelithiasis, and increased ALT and AST, and direct and total bilirubin.
The selected articles were published between 2015 and 2021, with six studying oral microbiota; four biliary; three intestinal per fecal sample, and two per duodenal samples (tissue or fluid). Mostly, they compare a controlled microbiota and a patient previously diagnosed with pancreatic or biliary cancer. Selected reviews include countries: Canada, China, South Korea, United States, Finland, Israel, France, and Italy.
The selected studies mostly analyzed cases of PDAC (13 studies), followed by CCA,4 Gallbladder Carcinoma (GBC),1 each study being possible to include one or more types of neoplasm of the biliopancreatic tract. Participants in each of the studies were separated into groups for microbiome analysis. The case groups formed were according to the type of cancer of the patients (PDAC and CCA, for example) or, as in Riquelme E. et al.,26 survival time of the patient with the disease. Controls were divided into healthy or benign disease patients (calculi in bile ducts and Intraductal Papillary Mucinous Neoplasm ‒ IPMN, for example). The same study may have presented more than one case or control groups, such as Sun, H. et al.,32 who had one healthy control and one with benign conditions.
Of the 11 selected studies, 9 used 16s rRNA as an analysis method to characterize the specific composition of the microbiota. On the other hand, Serra, N. et al.27 and Di Carlo, P. et al.28 analyzed the biliary microbiota using the BD Phoenix System. In addition, Di Carlo used the Vitek-2 system together.
In studies in which the oral microbiota was analyzed, Vogtmann, E. et al.29 has as control microbiota a predominance of the genus Haemophilus (from the phylum Proteobacteria), whose presence would decrease the chance of biliopancreatic neoplasia (OR = 0.95), while, for the case group, there was an increase in the families Lachnospiraceae G7, Bacteroidaceae, Staphylococcaceae and Enterobacteriaceae, but does not specify the genus. Olson, S.H., et al.33 cites the presence of the genus Neisseria and Haemophilus (both from the phylum Proteobacteria) in the control group and the genus Streptococcus (phylum Firmicutes) in the case group. In both cases, the disease was PDAC.
The control cases of Fan, X. et al.,22 Wei, AL. et al.34 and Torres, P.J. et al.,23 diverge in relation to the results of the analyses of the oral microbiota. Fan, X. et al. associate the low risk of PDAC with the presence of bacteria of the genus Leptotrichia (phylum Fusobacteria) (95% CI 0.89 to 0.99; OR = 0.87 and 95% CI 0.79 to 0.95) and high risk with the presence of Porphyromonas (phylum Bacteroidetes) (OR for presence vs. absence = 1.60 and 95% CI 1.15 to 2.22; OR = 2.20) and Aggregatibacter actinomycetemcomitans (phylum Proteobacteria) (OR = 2.20 and 95% CI 1.16 to 4.18). Torres, P.J. et al. and Wei, AL. et al., however, reported a decrease in these same bacteria genera in patients with PDAC, Porphyromonas and Neisseria, and an increase in Leptotrichia. Wei, AL. et al. also found an increase in Streptococcus.
Sun, H. et al.,32 found the genus Neisseria among the most abundant in the oral microbiome mainly in controls, but also in cases of PDAC, in alignment with Wei, AL. et al. and Torres, P.J. et al. On the other hand, Wei, AL. et al. associates Veillonella with low risk for PDAC, Sun, H. et al. found this genus as one of the most prevalent in cases of the disease.
In the analysis of the biliary microbiome, two cross-sectional studies and one cohort reported an increase in Escherichia (phylum Proteobacteria, family Enterobacteriaceae) in patients with CCA. Serra, N. et al.,27 and Di Carlo, P. et al.28 found an increase in Pseudomonas (phylum Proteobacteria, family Pseudomonadaceae), while Chen, B. et al.,25 found Klebsiella (phylum Proteobacteria, family Enterobacteriaceae), in addition to Faecalibacterium, Okibacterium and Corynebacterium. The case and control groups (composed of patients with choledocholithiasis) by Chen, B. et al., showed an increase in Escherichia/Shigella and Klebsiella, and only the case group, Halomonas, Streptococcus and Enterococcus. The control case of Saab, M. et al. diverged from the other three with bile analysis, with the CCA group presenting the genera Streptococcus, Pyramidobacter, and Bacteroidetes as more abundant compared to the controls, with no congruence with the other studies.
Di Carlo, P. et al.,28 and Serra, N. et al.27 analyzed the biliary microbiome in patients with PDAC. The first reported Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa as more frequent in patients, in contrast to the second, which, despite having found Klebsiella spp as a positive predictor, showed Escherichia spp and Pseudomonas spp as negative predictors. In addition, Serra, N. et al. reported Pseudomonas spp as a positive predictor for GBC.
The four articles that analyze the intestinal microbiota point to Firmicutes as among the most frequently found phyla, in cases and controls. In two of them, Proteobacteria also appeared in both groups. Q.-X. Mei et al.,30 when comparing the duodenal microbiota of patients with PDAC with healthy controls, place the genera Acinetobacter, Aquabacterium, Rahnella, Delftia, Sphingobium, Massilia, Oceanobacillus, Deinococcus as more abundant in the first group than in the second, while Escherichia, Shigella, Pseudomonas, Enhydrobacter, Porphyromonas, Paenibacillus were less abundant. For Kohi, S., et al.,35 however, Bifidobacterium, Enterococcus, Clostridium, Escherichia, Shiguella, and Fusobacterium were the most abundant genera in PDAC when compared to a benign disease or healthy controls, even though it was also an analysis of the duodenal flora.
Riquelme E. et al.,26 a cohort, compared the intestinal microbiota between patients with short (STS) and long (LTS) survival after the diagnosis of PDAC, obtaining a result that in LTS patients there is a predominance of the phyla Proteobacteria (genus Pseudoxanthomonas) and Actinobacteria (genera Streptomyces and Saccharopolyspora), in addition to the presence of Bacillus clausii. In STS patients, however, there were no predominant genera, but classes: Clostridia and Bacteroidea, contrary to Kohi S., et al.,35 who found, for these, the Fusobacterium, Rothia and Neisseria genera as the most prevalent and belonging to classes that differ from those mentioned above by Riquelme E. et al. Half et al.31 and Ren et al.24 show agreement at the family level regarding the increase of Veillonellaceae in PDAC and at the genus level regarding the greater presence of Clostridium in controls compared to cases. Otherwise, there were no findings common to these studies.
DiscussionTo reduce the mortality of pancreatic and biliary tract cancer, it is important to have methods that help in the early diagnosis and intervention of the disease. For this, studies relating to microbiota, whether fecal, biliary, or oral, with the incidence of PDAC and CCA have great importance in the academic world. This systematic review analyzed 15 articles that assess the microbiota of patients with cancers of the biliopancreatic, comparing it or not with that of controls.
About 86.67% of the studies use 16s rRNA as a sequencing method to assess the composition of the microbiota. This method is useful when there is no basic knowledge about the possible findings of the analysis, in addition to being a well-known and lower-cost method compared to other techniques. Despite this, the method is not the most suitable for detecting strains for epidemiological purposes or as a specific virulence factor.36 The other method used to approach the microbial composition was BD Phoenix System, which is culture-dependent, assessing only pathological bacteria. Considering this, a proper comparison between studies with different analysis methods is not possible, for the intrinsic selection bias caused by the distinct perspective of which one of them. Another factor that should be taken into account regarding the use of 16s rRNA sequencing is the no specificity of the method to any particular group, without restricting the taxonomic classification to be used by the researcher. One of the greatest difficulties encountered in the systematic review was the heterogeneity of the presentation of results in relation to taxonomic groups. For example, Fan, X. et al.22 reports the most frequently found species, while Vogtmann, E. et al.29 cites a genus and some families present in greater abundance, making it difficult to compare them.
To reduce the differences between the studies and standardize them, in order to make comparison possible, only phylum, family, and gender were included in the tables present in the results, and class, when the previous ones had not been made available by the author. It was necessary to research and classify the proposed taxonomic phyla (phylum, family, and genus), which was not possible in those studies that selected broader categories. Considering the importance of comparing studies, these should place more than one classification in the microbiota found or it should be agreed that studies on microbiota and its possible pathogenicity always select the same classifications, for example, family or genus.
There was also great variety in the ways to expose the constitution of the microbiome obtained after the analyses. Some articles, such as Di Carlo, P. et al.,28 only indicate which strains are most prevalent in each group, while others, such as Olson, S. H. et al.,33 do not describe the samples individually, but only the differences between groups. This makes it difficult and sometimes impossible to understand the actual composition of each sample and establish a pattern considered healthy and another characteristic of each disease studied. Despite this, there are articles in which this exposition was complete, describing both the composition of the samples by group and between different groups, as in Q.-X. Mei et al.,30 is a good model for future articles addressing this topic.
Another point to be highlighted is the difficulty in comparing the results of case-control studies, which diverged in terms of the phyla and genera found. This can be attributed to the different nationalities of the studies, such as Half, E et. al.,31 who is Israeli, and of Ren Z. et. al.,24 who is Chinese. The geographic difference is also reflected in lifestyle habits and genomic factors. Furthermore, these studies also differ regarding the specification of comorbidities and the inclusion of patients with laboratory alterations, which were carried out only by Half, E. et al. In this study, healthy people with Non-Alcoholic Fatty Liver Disease (NAFLD) were included, with comorbidities such as diabetes mellitus (13%), systemic arterial hypertension (50%) and dyslipidemia (88%), factors that can influence the composition of the microbiome.37 Thus, it is not possible to determine the effects of this difference in the composition of the fecal microbiota.
Although there are main phyla present in the intestine (Bacteroidetes and Firmicutes),38 the collective microflora is composed of more than 35,000 bacterial species39 and it is difficult to determine a static control composition, since even primary pathogens that inhabit the human intestine, in low incidence and in symbiosis, are referred to as healthy.40 Thus, it was not possible to determine what is, and if there really is, a microbiota to be used as a control.
In the studies by Ren, Z et al.24 and Mei, Q-X et al.,30 the phylum Proteobacteria was found in abundance in cases and controls. Kohi, S. et al.35 also found this for controls but diverged from Hollister, B et al.,40 who found little abundance of this phylum in healthy individuals and an increase in cases of gastrointestinal tract disease. Furthermore, Hollister, B et al. proposed Streptococcus as the main genus in the non-diseased duodenum, while Mei Q-X et al. does not mention it and Kohi S et al. found an increase in this genus only in proton pump inhibitors users. These disagreements reinforce the need for additional studies to determine the composition of the human microbiota in its various sites, health conditions, and interfering factors.
With regard to the comparison of the oral microbiota, Fan, X. et al.,22 from New York, cited the presence of bacteria of the genus Leptotrichia as low risk for PDAC and Porphyromonas and Aggregatibacter actinomycetemcomitans as high risk. On the other hand, Torres, P.J. et al.,23 whose study was conducted in San Diego, concluded that the presence of Porphyromonas and Neisseria is linked to low risk of PDAC while Leptotrichia is linked to high risk. Both studies took place in the same country with similar populations (ethnicity, risk factors, age), which would tend to reduce the difference between results in the microbiota found. Despite this, their conclusions were contradictory, while Wei AL et al.,34 a Chinese study, showed agreement with Torres, P.J. et al., probably due to different sample collection methodologies, in which Fan, X. et al. collected saliva with mouthwash, while the other two collected the material without the use of other liquids. This reinforces the difficulty of comparing microbiota.
On the other hand, Vogtmann, E. et al.39 and Olson, S.H., et al.33 agree in citing the presence of the genus Haemophilus and its higher taxonomic levels as protective, despite approaching different populations, the first from the northeast of the USA and the second from Iran.
Bile was the analyzed sample that had the greatest agreement among the results. Serra, N. et al.27 and Di Carlo, P. et al.28 describe a positive correlation between the presence of Klebsiella and PDAC, but they diverge as to the role of the genera Escherichia and Pseudomonas, as the first classified them as negative predictors for the disease and the second as the most common genera. But this difference can be explained by the type of analysis carried out by Serra N. et al., who compared three different diseases (PDAC, CCA and GBC) and, based on this comparison, arrived at these results. But the similarity may be explained by the BD Phoenix System used for analysis, which restricts the searched composition to the clinically significant bacteria. This may also explain the difference between the latter and Chen. B et al.,25 for which the presence of Klebsiella is related to the appearance of CCA, but the investigation was made using 16S rRNA for sequencing.
The great disagreement regarding the microbiota among the selected articles can be explained by the various factors that influence it. Among them are age, hygiene, life habits, diet, and other external factors,41 therefore, it is possible to claim that there will always be a difference in the microbiome, especially in very different cultures. Establishing a consensus on the taxonomic description and obtaining samples is essential to allow comparison between results. For future studies that seek to assess the impact of cancer on the composition of the microbiome, the material should be collected at two different times: one before and one after the suspicion and diagnosis.
After analyzing the selected articles and given the limitations described, it is not possible to state that any microorganism can be related to pathogenicity, colonization, or used as screening for patients with PDAC or CCA.
In an attempt to more accurately characterize the biliary microbiota related to PDAC, there is work being carried out by the same team of this review that aims to analyze the difference between the microbiota of healthy people compared to patients with hepatobiliopancreatic diseases.42
ConclusionThere was great disagreement in the characterization of both the microbiota of patients with benign diseases and patients with cancer of the biliopancreatic tract. The literature is still more focused on the study of the intestinal microbiota, with comparisons being made between healthy patients and those with PDAC. Thus, studies that analyze the microbiome of other sites, such as biliary and pancreatic, or its possible alterations in diseases such as CCA, are still scarce, making it difficult to adequately assess the data in this regard. In addition, the composition of the microbiota is greatly influenced by lifestyle habits and comorbidities, and it is questioned whether there really is a microbiota to be defined as normal. Due to these factors, it was not possible to find any specific marker or to associate any genus of microbiota bacteria with PDAC or CCA. More studies are needed not only to determine cancer-associated virulence factors but also to characterize healthy and pathogenic microbiota.
AbbreviationsPDAC, Pancreatic Ductal Adenocarcinoma; CCA, Cholangiocarcinoma; NFKB, Nuclear Factor Kappa B; VEGF, Vascular Endothelial Growth Factor; PRISMA-P, Preferred Reports for Systematic Reviews and Protocol Meta-Analysis; PROSPERO, Prospective Register of Systematic Reviews; PICO, Patient or Problem, Interest, Control or Comparison, Outcome; IPMN, Intraductal Papillary Mucinous Neoplasms; PBD, Pancreas and Biliary tract Disease; LTS, Long Term Survival; STS, Short-Term Survival; HTS, High-Throughput Sequencing; ERCP, Endoscopic Retrograde Cholangiopancreatography; M, Male; F, Female; CBD, Common Bile Duct; RC, Recurrent Choledocholithiasis; PC, Pancreatic Cancer; PCL, Precancerous Lesions; NAFLD, Non-Alcoholic Fat Liver Disease; GBC, Gallbladder Carcinoma; DM, Diabetes Mellitus; AST, Aspartate Aminotransferase; ALT, Alanine Aminotransferase; Tbil, Total Bilirubin; Dbil, Direct Bilirubin; Scr, Serum Creatinine; WBC, White Blood Cell; NE, Neutrophilic Granulocyte.
Ethics approval and consent to participateNot applicable.
Consent to publishNot applicable.
Availability of data and materialsAll data produced and obtained is available within the manuscript.
Authors' contributionsAM had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Concept and design: VCM, FSN, MOS, AM. Acquisition, analysis, or interpretation of data: VCM, FSN, MOS, JVT, LZP, WAFM, VSC, WTH, LRI, MSK, LACDA, AM, WA. Drafting of the manuscript: LACD, VCM, FSN, MOS, WAFM. Critical revision of the manuscript for important intellectual content: AM, LRI. Statistical analysis: MOS, JVT, LZP, VSC, MBT. Administrative, technical, or material support: VCM, WAFM, WTH, LRI. Supervision: AM, LACDA, WA. All authors have read and approved the manuscript.
FundingNo funding.
The authors are thankful to Justin Axel-Berg for the English corrections, Rossana V. Mendoza López for statistical analysis and Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), Processos no. 2020/08330-3 and no. 2020/08328-9.