In endemic areas, a small proportion of individuals infected with dengue virus develop dengue hemorrhagic fever (DHF) suggesting that there may be host specific resistance factors playing an important role. This work describes the frequency of HLA class I and class II alleles in patients with dengue and the relationship with the clinical manifestations of the disease. The analysis of the frequency of HLA specificities in the dengue patients revealed reduced frequencies of B*15, B*49, DRB1*02 and DRB1*03 and increased frequencies of B*57 and DRB1*15 compared with controls. When the patients were grouped and compared according to disease severity, an association with enhanced susceptibility to dengue fever (DF) in patients with B*57, an association with reduced susceptibility to DHF in patients with A*03, and an association with enhanced susceptibility to DHF in patients with B*40 was observed. Although the associations revealed in this study come from a very small case-control population and that after correction for multiple testing only the association with DRB1*15 is maintained, the data suggest that the HLA alleles can possibly play a role in the susceptibility and/or resistance to dengue virus infection and development of DHF.
En áreas endémicas, una pequeña proporción de individuos infectados con el virus dengue desarrollan la fiebre hemorrágica del dengue (FHD), sugiriendo la existencia de factores de resistencia que pudiesen estar jugando un papel importante en el huésped. Este trabajo describe la frecuencia de alelos HLA clase I y II en pacientes con dengue y su relación con las manifestaciones clínicas de la enfermedad. El análisis de la frecuencia de especificidades HLA en los pacientes con dengue mostró frecuencias disminuidas de los alelos B*15, B*49, DRB1*02 y DRB1*03 y frecuencias incrementadas de los alelos B*57 y DRB1*15 comparado con los controles. Cuando los pacientes fueron agrupados y comparados de acuerdo a la severidad de la enfermedad, se observó una susceptibilidad incrementada a la fiebre del dengue clásico (FD) en pacientes con B*57, una susceptibilidad disminuida al desarrollo de la FHD en pacientes A*03 y una susceptibilidad incrementada al desarrollo de la FHD en pacientes con B*40. Aunque las asociaciones observadas proceden de un estudio poblacional caso-control relativamente pequeño y que después de la corrección por múltiples comparaciones se mantuvo únicamente la asociación con DRB1*15, los datos confirman que posiblemente los alelos HLA pueden jugar un papel en la susceptibilidad y/o resistencia a la infección por virus dengue y al desarrollo de la FHD.
Dengue virus infections are an important cause of morbidity and mortality in tropical and subtropical areas(1). In humans, the infection can be asymptomatic or can result in a variable range of clinical syndromes, including classical dengue fever (DF) and the more severe form of disease known as dengue hemorrhagic fever (DHF)(2). In Venezuela, the dengue virus serotypes 1, 2 and 4 are endemic, they circulate simultaneously throughout the year in the largest cities(3), however severe epidemics of dengue or DHF in Venezuela have been shown to be the result of the introduction of dengue virus serotype 3 in 2001(4).
Several hypotheses have been postulated to explain the pathogenesis of severe dengue disease, with immunopathogenesis as the most supported model. It has been suggested that polymorphisms in the cytokine genes may play an important role in the pathogenesis of DHF(5). However, other immune response-related genes (e.g. HLA genes), as well as non-genetic factors such as dengue virus serotype and host viral burden, must also be considered.
The association of DHF with HLA antigens has been reported in ethnically and geographically distinct populations(6–12). In this study we have analyzed the frequency of HLA class I (−A, −B and −C) and class II (−DRB1) polymorphisms in Venezuelan patients with DF and DHF in order to understand the relative contribution of these genes to disease pathogenesis.
MATERIALS AND METHODSPatient and control Populations.Whole blood was collected from 77 unrelated patients (mean age 27, range 1-82 years). Using all available clinical and laboratory data, 73 of the 77 patients were classified into two different clinical groups: 43 with DF, and 28 with DHF. Six patients, albeit infected with dengue virus, were not clinically classified.
All patients were ethnically mixed Venezuelans, i.e. born in Venezuela and descended mainly from Spanish Europeans, autochthonous inhabitants, and West Africans forced to come during colonial times. A previous study using genetic markers has shown that the origin of genes carried by the central urban Venezuelan population is approximately 60.4% European, 23.5% Amerindian and 16.1% African(13).
Dengue viral infection was confirmed for each patient during illness by the detection of antibodies (IgG and IgM) specific against the dengue virus (ACON Laboratories, Inc. 4108 Sorrento Valley Boulevard. San Diego, USA); these humoral responses discard other flaviviruses that are endemic in South America. However, antibody concentrations in serum were not registered in the clinical history of the patients. The clinical diagnosis was confirmed in accordance with the World Health Organization (WHO) criteria (2), using all available clinical and laboratory data. They were clinically classified as patients with dengue fever (DF), an acute febrile illness with two or more of the following manifestations: headache, retro-orbital pain, myalgias, arthralgias, rash, mild hemorrhagic manifestations, and patients with DHF (fever, severe hemorrhagic manifestations, thrombocytopenia at or below 100.000/mm3, elevated haematocrit). It was not possible as part of this study to determine the serotype of the dengue virus.
One hundred-twenty seven unrelated healthy subjects of similar ethnic background and without signs of infection were tested as controls. A local ethics committee approved the research protocol.
HLA TypingGenomic DNA was extracted from blood samples using a modified salting-out procedure(14). HLA typing was carried out by polymerase chain reaction-sequence-specific oligonucleotide reverse dot blot using the Dynal RELI™ SSO HLA-A, -B, -C and DRB test kits. HLA-A, HLA-B, HLA-C, HLA-DRB1 allelic groups were defined in patients and controls.
The HLA class I alleles were evaluated in 127 healthy individuals and 77 patients with dengue, while the HLADRB1 polymorphism was determined in 97 controls and 77 patients.
Statistical AnalysisAllele and genotype frequencies were determined by direct counting. The statistical significance of allele frequency differences between patients and controls was estimated by Fisher's exact test using 2x2 contingency tables; p values were corrected multiplying by the number of comparisons made (Bonferroni correction) and were considered significant when p <0.05. Relative risks with corresponding 95% confidence intervals (95% CI) were calculated as odds ratios (OR) according to Woolf's formula(15) or by the modified method described by Haldane(16) when one element of the equation was zero. The OR is used to estimate risk in casecontrol studies in which the relative risk calculations is not appropriate. An OR <1 indicates protection, whereas an OR >1 indicates an increased risk.
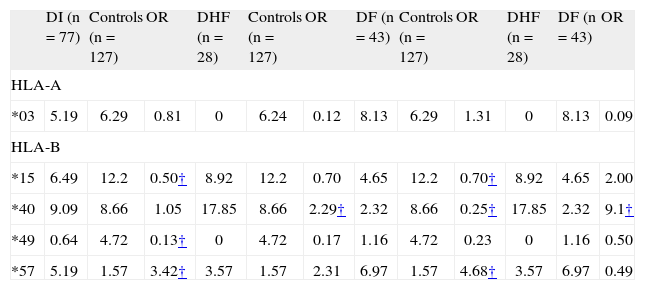
RESULTSTable I shows the distribution of selected HLA class I allele frequencies in control subjects and patients with dengue infection (DF and DHF). The analysis of HLA-class I profiles in the patient and control groups showed three significant associations. HLA-B*15 (5 patients with *1503, 3 patients with *1501, 1 patient with *1504 and 1 patient with *1509) and B*49 were significantly reduced in frequency in patients with dengue infection compared with healthy individuals. In contrast, HLA-B*57 was significantly increased in patients with dengue when compared with the controls. No significant differences in HLA-A and HLA-C were observed between the patients and the controls.
HLA-class I specificities with frequency differences among patients with dengue infection (DI; entire group) and control subjects, and patients with dengue fever (DF) versus patients with dengue hemorrhagic fever (DHF). Results are presented as phenotype frequency percent
| DI (n = 77) | Controls (n = 127) | OR | DHF (n =28) | Controls (n = 127) | OR | DF (n =43) | Controls (n = 127) | OR | DHF (n =28) | DF (n = 43) | OR | |
| HLA-A | ||||||||||||
| *03 | 5.19 | 6.29 | 0.81 | 0 | 6.24 | 0.12 | 8.13 | 6.29 | 1.31 | 0 | 8.13 | 0.09 |
| HLA-B | ||||||||||||
| *15 | 6.49 | 12.2 | 0.50† | 8.92 | 12.2 | 0.70 | 4.65 | 12.2 | 0.70† | 8.92 | 4.65 | 2.00 |
| *40 | 9.09 | 8.66 | 1.05 | 17.85 | 8.66 | 2.29† | 2.32 | 8.66 | 0.25† | 17.85 | 2.32 | 9.1† |
| *49 | 0.64 | 4.72 | 0.13† | 0 | 4.72 | 0.17 | 1.16 | 4.72 | 0.23 | 0 | 1.16 | 0.50 |
| *57 | 5.19 | 1.57 | 3.42† | 3.57 | 1.57 | 2.31 | 6.97 | 1.57 | 4.68† | 3.57 | 6.97 | 0.49 |
Odds ratio 95% confidence interval;
B*15 ↑ healthy vs. DI (protection)A*3 ↑ healthy vs. DHF (protection)B*57 ↑ DF vs. healthy (susceptibility, OR: 4).
B*49 ↑ healthy vs. DI (protection)B*40 ↑ healthy vs. DF (protection)B*40 ↑ DHF vs. DF (susceptibility, OR: 9).
B*15 ↑ healthy vs. DF (protection)B*57 ↑ DI vs. healthy (susceptibility, OR: 3)B*40 ↑ DHF vs. healthy (susceptibility, OR: 2).
HLA-class I profiles in the patients with DF and DHF, showed significant disease associations. HLA-B*40 is present at increased frequency among patients with hemorrhagic manifestations compared to healthy individuals (17.85 vs. 8.66%; OR = 2.29; 95% CI: 1.01-5.16; p= 0.035) and patients with DF (17.85 vs. 2.32%, respectively; OR = 9.1; 95%CI: 1.91-43.45; p= 0.016), and B*57 is present at increased frequency among patients with the clinically less severe DF compared to healthy individuals (6.97 vs. 1.57%, respectively; OR = 4.68; 95% CI: 1.29-17.02; p= 0.014). Finally, A*03 is absent in patients with DHF compared with the control group (0% vs. 6.29%, respectively; OR = 0.12; 95% CI: 0.007-2.16; p= 0.04). However, all of these HLA class I frequency differences lost significance after Bonferroni correction.
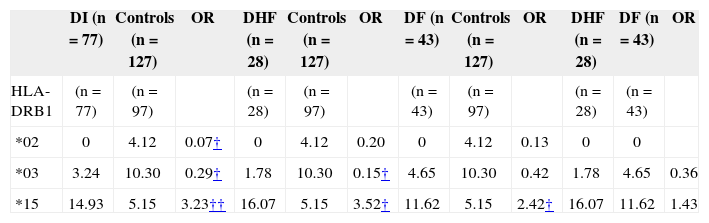
Table II shows the distribution of selected HLA-DRB1* types in dengue patients and controls. When the frequency of HLA-DRB1 alleles in the patients with DF and controls (only 97 healthy individuals were tested for HLA-DRB1) were compared, a significant decrease of DRB1*02 and DRB1*03 in the whole series of dengue patients was observed; however, these probability values lost significance after correction (pc). In contrast, the patients with DF showed a significant increase of the DRB1*15 compared with healthy individuals (14.93 vs. 5.15%, respectively; OR = 3.23; 95% CI: 1.48-7.01; p= 0.0018; pc=0.0252), which remained significant after stratifying by syndromes, DHF (16.07 vs. 5.15%, respectively; OR = 3.52; 95% CI: 1.35-9.16; p=0.007; pc=ns) and DF (11.62 vs. 5.15%, respectively; OR = 2.42; 95% CI: 0.96-6.05; p= 0.045; pc=ns).
HLA-DRB1 specificities with frequency differences among patients with dengue infection (DI; entire group) and control subjects, patients with dengue fever (DF) versus control and patients with dengue hemorrhagic fever (DHF) versus healthy individuals. Results are presented as phenotype frequency percent
| DI (n = 77) | Controls (n = 127) | OR | DHF (n = 28) | Controls (n = 127) | OR | DF (n = 43) | Controls (n = 127) | OR | DHF (n = 28) | DF (n = 43) | OR | |
| HLA-DRB1 | (n = 77) | (n = 97) | (n = 28) | (n = 97) | (n = 43) | (n = 97) | (n = 28) | (n = 43) | ||||
| *02 | 0 | 4.12 | 0.07† | 0 | 4.12 | 0.20 | 0 | 4.12 | 0.13 | 0 | 0 | |
| *03 | 3.24 | 10.30 | 0.29† | 1.78 | 10.30 | 0.15† | 4.65 | 10.30 | 0.42 | 1.78 | 4.65 | 0.36 |
| *15 | 14.93 | 5.15 | 3.23†† | 16.07 | 5.15 | 3.52† | 11.62 | 5.15 | 2.42† | 16.07 | 11.62 | 1.43 |
Odds ratio 95% confidence interval;
*02 ↑ healthy vs. DI (protection)*03 ↑ healthy vs. DHF (protection)*15 ↑ DF vs. healthy (susceptibility, OR: 2).
*03 ↑ healthy vs. DI (protection)*15 ↑ DI vs. healthy (susceptibility, OR: 3)*15 ↑ DHF vs. healthy (susceptibility, OR: 3)
Dengue virus infection has emerged as one of the most important arthropod-borne diseases. It is well documented that viral burden and infected cell mass determine the clinical severity of dengue infections; however, the ability of the host to recognize and process antigens to produce antibodies or the cellular immune response during dengue infection could be under genetic control(4). Loke et al. showed that the HLA class I region polymorphism, particularly of the HLA-A gene, is a significant determinant of genetic susceptibility to DHF(17). Stephens et al. reported different HLA class I alleles associated with the severity of clinical disease in secondary dengue virus infection, suggesting that classical HLA class I gene products play a crucial role in determining the outcome after exposure to different dengue virus serotypes, in previously exposed and immunologically primed individuals (11). More recently, Lan et al.(12), reported that their study about HLA class I in dengue disease reproduced a previous Vietnamese study(17), in which HLAA*24 is associated with DFH and DSS (dengue shock syndrome), and showed that the frequency of A*24 alleles with histidine at codon 70 (A*2402, 2403, 2410) among DHF and DSS patients were significantly higher than that in the control group. Although evidence to date suggests that HLA class I genes correlate with both enhanced and decreased susceptibility to DHF, a study performed in Mexican patients with dengue virus infection revealed that the HLA-DRB1 locus may also protect patients from developing DHF. In this study, the authors suggest that immunological determinants of protein E (the envelope protein of the virus) are probably processed and presented by class II MHC antigens and that the HLA-DRB1*04 molecule may present these viral antigens to CD4+ lymphocytes leading to an effective immune response and consequently protection from DHF(9). A protective effect of HLA-DRB1*0901 against DSS development from DHF in Southern Vietnam patients with DEN-2 infection is a new finding providing evidence for HLA class II control of dengue severity(12). Furthermore, a positive association of HLA-DQ1 and susceptibility to classical dengue fever in a white Southern Brazilian population has been reported(10).
The present study is the first to examine a complete set of HLA polymorphisms and to determine all class I and class II alleles in infected Venezuelan individuals using molecular methods. The preliminary results of our study among patients with dengue virus infection have shown several positive and negative associations, suggesting either predisposing or protective effects of MHC genes on the development of the infection. Analysis of HLA alleles in the group of healthy individuals revealed increased frequencies of B*15, B*49, DRB1*02 and DRB1*03 compared with dengue patients (DF+DHF), suggesting that these allele groups could be associated with diminished susceptibility to the infection. In contrast, B*57 and DRB1*15 showed an increased frequency in patients compared with healthy individuals, suggesting that these alleles could be associated with susceptibility to the infection.
When the patients were grouped according to disease severity, an association with diminished susceptibility to DHF in patients with A*03, an association with enhanced susceptibility to DHF in patients with B*40, and a positive association of B*57 with DF was observed. It is important to emphasize that our results do not support the associations reported in studies performed in other populations, but support the notion that polymorphism in the HLA class I region is a significant determinant of genetic susceptibility to DHF, suggesting an important role for CD8+ T cell responses in the development of dengue hemorrhagic fever(17).
The results suggest that polymorphism in the HLA class I and II genes may be associated with genetically diminished and enhanced susceptibility to the development of dengue virus infection and with disease severity (DHF) in the studied population. It is possible that the pathogenesis of dengue hemorrhagic fever may involve different HLA specificities together with other non-HLA gene families. Thus, variations in the vitamin D receptor and FCGRIIA genes(18), as well as variations in cytokines(5) and CTLA-4 genes(19) might confer susceptibility to DHF. Although we have previously reported in the same DHF patients a significant increase of the TNF-α308A(5), an increase of DRB1*03 is not evident among those carrying the TNF-α308A allele, reflecting the heterogeneity of Venezuelans due to migrations from other continents and within the continent(20). If further studies in a larger series of patients confirm the results here obtained, individuals carrying HLA-A*03 in our population might be protected to develop DHF, while those possessing B*40 will have more chances to develop the more severe form of the disease, conferring susceptibility among dengue seropositive individuals (OR= 9) and among healthy individuals (OR=2). Likewise, individuals with HLA-DRB1*15 might be more susceptible to the infection with dengue virus (OR= 3, pc<0.05).
The ability to assess several polymorphisms throughout the human genome may provide insights into possible mechanisms of pathogenesis and protection in dengue(18). Meanwhile, the genetic background of the host, the viremia titer, the antibody response pattern and virus serotype seem to be correlated with disease severity.
ACKNOWLEDGMENTSThis research was financially supported by FONACIT grant S1-2002000504. Our gratitude to the patients who participated in the study and to Ingrid Márquez and Federico Naranjo for help in the collection of samples.
CONFLICT OF INTERESTThe authors declare no financial conflict of interest.