Helicobacter pylori adheres to various components of the human saliva. Therefore, the objective of this research was to simultaneously detect H. pylori in saliva and in gastric biopsy, and to determine the agreement between the vacA genotypes in both saliva and gastric biopsy.
Materials and methodsA total of 162 patients with chronic gastritis and 34 with gastric ulcer were studied, and saliva and biopsy samples were collected from each patient. H. pylori DNA was detected by conventional PCR and nested PCR was used for vacA genotyping.
ResultsIn 24% of the patients (47/196) H. pylori DNA was found in saliva and in biopsy; 52.5% (103/196) were salivanegative/biopsypositive and 6.6% (13/196) were salivapositive/biopsynegative. In either or both H. pylori vacAs1m1 or s1m2 genotypes were detected in saliva in 41.5% of the patients with chronic gastritis. Forty-seven percent had >1 genotype, and the s1m1/s1m2 combination was found in 36% of them. H. pylori vacAs1m1 and s1m2 were also found in the saliva and biopsy of patients with gastric ulcer. The genotypes found in saliva and biopsy of the same patient had 51.1% agreement. In 27.6% of the 47 patients salivapositive/biopsypositive two genotypes were found in saliva, and one or both in the stomach.
ConclusionsThe s1m1/s1m2 genotypes, alone or together, are found simultaneously in saliva and gastric biopsy of the same patient. These results suggest that H. pylori reaches the oral cavity by various ways, and that saliva can be the transmitting and re-infecting vector.
Helicobacter pylori (H. pylori) se adhiere a diversos componentes de la saliva humana, por ello, el objetivo de esta investigación fue detectar H. pylori en saliva y biopsia gástrica y determinar la concordancia entre los genotipos vacA encontrados en saliva y estómago del mismo paciente.
Material y métodosSe estudiaron 162 pacientes con gastritis crónica y 34 con úlcera gástrica. De cada paciente se obtuvo una muestra de saliva y biopsia gástrica. El ADN de H. pylori se detectó por PCR convencional y la genotipificación de vacA se hizo por PCR anidada.
ResultadosEn el 24% (47/196) de los pacientes se encontró ADN de H. pylori en saliva y biopsia; el 52,5% (103/196) fueron salivanegativos/biopsiapositivos y el 6,6% (13/196) resultaron salivapositivos/biopsianegativos. En 41,5% de los pacientes con gastritis crónica, se encontró H. pylori vacA s1m1, s1m2 o ambos en saliva. El 47,2% tenían>1 genotipo y en 36% de esos se encontró la combinación s1m1/s1m2. H. pylori vacA s1m1 y s1m2 también se encontró en saliva y biopsia de pacientes con úlcera. El acuerdo entre los genotipos encontrados en saliva y biopsia de los mismos pacientes fue del 51,1%. En el 27,6% de los 47 pacientes salivapositivos/biopsiapositivos se encontraron 2 genotipos en saliva, y uno o los 2 también se encontraron en estómago.
ConclusionesLos genotipos s1m1/s1m2 solos o coexistiendo se encuentran simultáneamente en saliva y biopsia de los mismos pacientes. Los resultados sugieren que H. pylori alcanza la cavidad oral por diversas vías y la saliva puede servir de vehículo para la transmisión y reinfección.
The prevalence of Helicobacter pylori infection varies with the geographic area and age of the population. In developing countries the prevalence in adults is 80% and in developed countries the prevalence is 20–50%.1,2 Torres et al. found a seroprevalence of 66.6% in Mexicans.3
The oral cavity can be a natural or a transient reservoir, source of stomach re-infection and person-to-person transmission. H. pylori has been isolated from saliva and dental plaque of patients with gastroduodenal diseases. The incidence is higher in children with infected parents, in African children whose mothers previously chew their food and in persons who share toothpicks.4–8
H. pylori adheres to sialylated or sulfated MUC5B salivary mucin by means of babA.9 Therefore, saliva can be the vector for pathogen transmission, and the source of infection and re-infection of the stomach after eradication therapy.4,6,9 The role of H. pylori in the oral cavity is still controversial and the detection rate of the bacterium in the mouth is 0–100% in healthy people and in patients with gastric disorders.2,4–8,10,11
The risk for developing gastric cancer is related to the vacA gene and other H. pylori virulence genes. The vacA gene, present in all H. pylori strains, is polymorphic and the toxin production levels depend on the genotype. H. pylori strains with vacA s1/m1 genotype produce high cytotoxin levels, the s1/m2 strains moderate levels and the s2/m2 strains produce little or no toxin.2,10 The VacA cytotoxin produced by genotypes vacA s1m1 or s1m2 of H. pylori causes injury in the gastric epithelium and stimulates an inflammatory process originating gastritis that can develop into chronic gastritis or gastric ulcer.2,12–16
The vacA gene is widely studied in gastric pathology but there are few reports on its presence in the oral cavity.2,17,18 It is not known if the bacterial allelotypes colonizing the mouth are the most frequently detected in the stomach of patients with gastritis, gastric ulcer or cancer. In Mexico there are no reports of the simultaneous detection of H. pylori vacA genotypes in the mouth and the stomach of patients with chronic gastritis and gastric ulcer. The genotyping of H. pylori in the mouth will reveal important data concerning the relationship of the mouth and stomach genotypes.
The main goal of this research was to determine simultaneously the prevalence of H. pylori and of vacA genotypes in the oral cavity and in the gastric mucosa of patients with chronic gastritis or with gastric ulcer and to establish if there is any agreement between the genotypes detected in the mouth and the stomach.
Materials and methodsPatientsA total of 196 patients with dyspepsia symptoms were subjected to digestive endoscopy at the Specialized Unit of Gastroenterology and Endoscopy in Chilpancingo, Guerrero, México. The included patients were not on anti-microbial treatment, and had not taken proton pump inhibitors or gastric pH neutralizers during the month previous to the endoscopic procedure. The patients with non-steroid anti-inflammatory treatment were excluded from the study. The patients or the parents of young children signed a letter of consent and the project was approved by the Ethics Committee of Guerrero Autonomous University.
Sample collectionBefore endoscopy, 2–3mL saliva samples were collected and placed in sterile propylene flasks containing 10mM, pH 8.0 Tris, 20mM, pH 8 EDTA and 5% SDS buffer. The samples were kept at −20°C until DNA extraction.
After an eight hour fast, the endoscopy was carried out with a video processor and a video gastroscope (Fujinon, Wayne, NJ, USA). An experienced gastroenterologist–endoscopist evaluated the endoscopy procedure. During endoscopy, two antral biopsies were taken from the stomach of each patient. In some cases the biopsy sections were taken from body and in patients with gastric ulcer, the biopsies were taken from ulcer margin. One of them was immediately placed in formaldehyde for the histopathology study and the other biopsy in the buffer already mentioned (10mM, pH 8.0 Tris, 20mM, pH 8 EDTA and 5% SDS) was kept at −20°C until H. pylori diagnosis was achieved. An experienced medical pathologist evaluated the biopsies. The histological diagnosis of patients was established using the criteria described in the classification update Sydney system.19
H. pylori detectionTotal genomic DNA was obtained from gastric biopsies, from saliva and from bacteria using phenol-chloroform-iso-amyl alcohol method after digestion with proteinase K.20H. pylori DNA was detected with HP16-219 5′-GCTAAGAGATCAGCCTATGTCC-3′ and HPGR16SR 5′-CAATCAGCGTCAGTAATGTTC-3′ oligonucleotides that amplify a segment of the rRNA 16S gene.16 The PCR reaction mix consisted of 150ng of total DNA from biopsy or 450ng from saliva DNA, 2.5mM MgCl2, 0.2mM dNTPs (Invitrogen, Carlsbad, CA, USA), 10pmol of each oligonucleotide and 1U of Platinum®Taq DNA Polymerase (Invitrogen, Carlsbad, CA, USA) in a 15μL final volume. The amplification program included a 5min 94°C cycle; 40 cycles at 94°C for 30s; at 55°C for 30s; at 72°C for 1min and one cycle at 72°C for 7min. The PCR-amplified products were analyzed by electrophoresis on 1.5% agarose gels. The gels were stained with ethidium bromide and examined under ultraviolet light. Samples were scored as positive when a band of 522bp could be detected in agarose gel. A patient was considered to be infected with H. pylori if the PCR test was positive.
DNA from strains of Staphylococcus aureus and Escherichia coli isolated from gastric biopsies, as well as from Campylobacter sp. was assayed with PCR 16S rDNA to confirm the specificity of the primers.
Genotyping of vacANested PCR was used for genotyping the vacA gene in H. pylori-positive samples with the primers reported by Koehler et al.21 In the first amplification round of the signal region the oligonucleotides used were: vacA1F 5′-CTGGT(C/T)TAAAGTCGCACCCTTTGTGC-3′ and vacA1R 5′-CAATGGCTGGAATGATCACGGTTGT(A/G)-3′; for the second round vacA2F 5′-CAAACACACCGCAAAATCAATCGCCC-3′ and vacA1R were employed. For the m1 allele detection in the first round the following primers were used: m1F1 5′-CAACAATCAAGGCACTATCAA(C/T)TA-3′, m1R1 5′-CCGCATGCTTTAATGTCATCAG-3′ and for the second round m1F2 5′-TGGTCCGAGGCGGG(A/C)AAGT-3′ and m1R2 5′-TCATCAGTATTTCGCACCACAC-3′21 were used. For the m2 allele the primers in the first run were: m2F1 5′-TTTGGAGC(C/T)CCAGGAAACATTG-3′ and m2R1 5′-C(C/T)ACACGCCCATCTTGGACAA-3′ and in the second run m2F2 (5′-ACCCTAAA(C/T)AGCAACGCAAGC-3′) and m2R2 (5′-GACAAAAAGATTCATCGTGCCTT-3′).The PCR for all these assays was: 700ng total DNA 1mM of MgCl2; 0.15mM of dNTPs (Invitrogen, Carlsbad, CA, USA), 10pmol of each nucleotide and 1U Platinum®Taq DNA Polymerase (Invitrogen, Carlsbad, CA, USA).
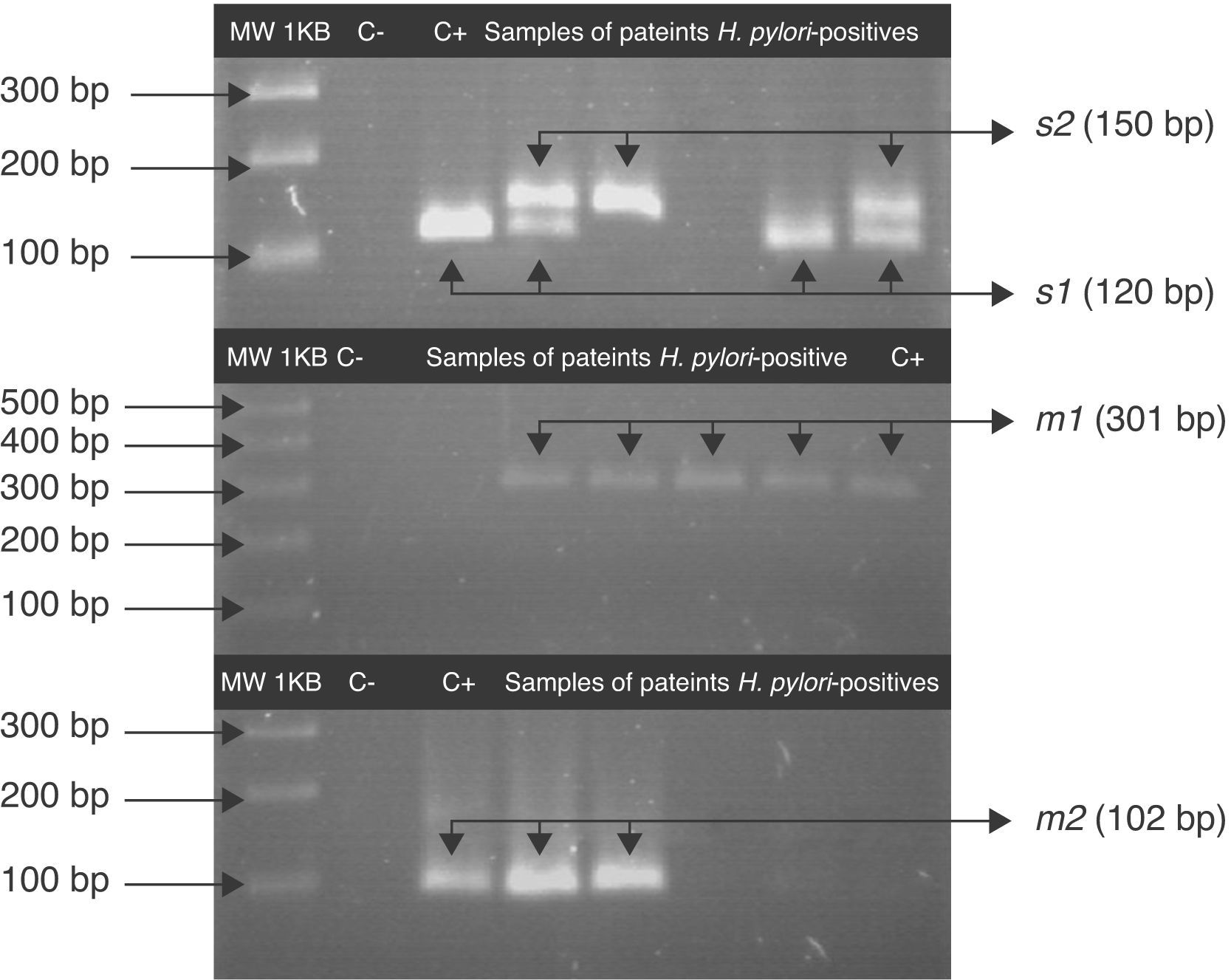
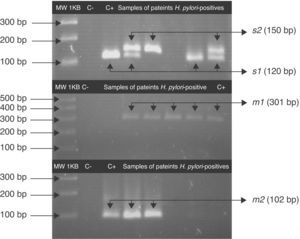
For the first amplification a 3min 95°C cycle was followed by 23 cycles: at 95°C for 40s; at 58°C for 40s at 72°C for 45s and a cycle at 72°C for 3min. For the second PCR 1μL of the product from the first reaction was the template and was amplified for 34 cycles. All PCRs were carried out in the above-mentioned conditions. The PCR products were analyzed by 3% agarose gel electrophoresis. A 120pb band corresponded to the s1 allele, a 150pb to the s2 allele and a 102pb band corresponded to m2. For the vacA m1 the agarose gel was 1.5% and a 301pb band was observed.
ATCC43504 strain was used as a positive control for H. pylori and for the vacA s1/m1 genotype and sterile deionized water was the negative control. For the m2 region the positive control was DNA from a gastric biopsy containing that allelotype. For all PCR a thermo Mastercycler Ep gradient (Eppendorf, Germany) was used.
Statistical analysisThe Stata V.10.0 computer statistical program was used. Absolute and relative frequencies of the qualitative variables were obtained. The kappa index was used to calculate the concordance of the presence of vacA genotypes of H. pylori in oral cavity and in gastric mucosal. The Chi square test or the exact Fisher's test was used to compare ratios of qualitative variables and to establish differences among groups. A p<0.05 value was considered significant.
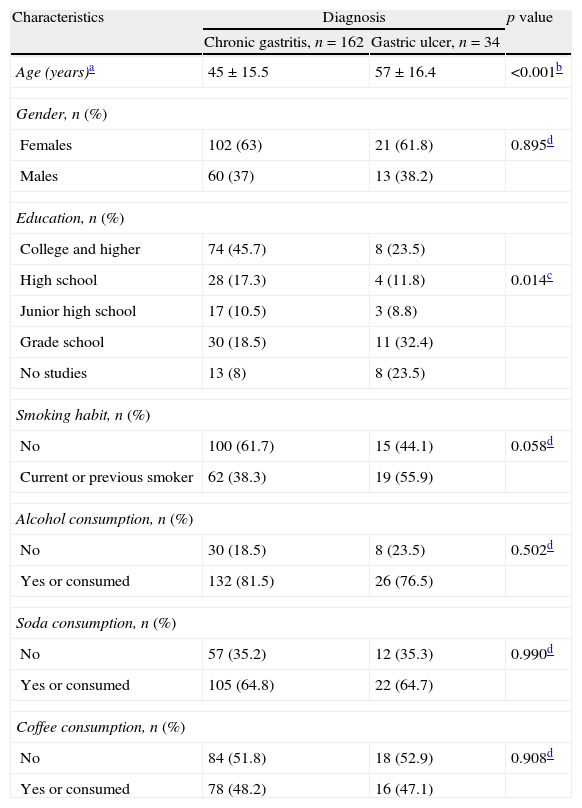
ResultsOf the 196 patients with dyspepsia, 162 (82%) had histopathological diagnosis of chronic gastritis and 34 (17%) of gastric ulcer. The mean age was 45 years old, range of 11–82 years for patients with gastritis. For gastric ulcer patients the mean age was 57 years old, range of 25–83. The age of patients with gastric ulcer was significantly more advanced than that of patients with chronic gastritis (p≤0.001). Female patients predominated (Table 1).
Socio-demographic characteristics of patients with chronic gastritis and with gastric ulcer.
| Characteristics | Diagnosis | p value | |
| Chronic gastritis, n=162 | Gastric ulcer, n=34 | ||
| Age (years)a | 45±15.5 | 57±16.4 | <0.001b |
| Gender, n (%) | |||
| Females | 102 (63) | 21 (61.8) | 0.895d |
| Males | 60 (37) | 13 (38.2) | |
| Education, n (%) | |||
| College and higher | 74 (45.7) | 8 (23.5) | |
| High school | 28 (17.3) | 4 (11.8) | 0.014c |
| Junior high school | 17 (10.5) | 3 (8.8) | |
| Grade school | 30 (18.5) | 11 (32.4) | |
| No studies | 13 (8) | 8 (23.5) | |
| Smoking habit, n (%) | |||
| No | 100 (61.7) | 15 (44.1) | 0.058d |
| Current or previous smoker | 62 (38.3) | 19 (55.9) | |
| Alcohol consumption, n (%) | |||
| No | 30 (18.5) | 8 (23.5) | 0.502d |
| Yes or consumed | 132 (81.5) | 26 (76.5) | |
| Soda consumption, n (%) | |||
| No | 57 (35.2) | 12 (35.3) | 0.990d |
| Yes or consumed | 105 (64.8) | 22 (64.7) | |
| Coffee consumption, n (%) | |||
| No | 84 (51.8) | 18 (52.9) | 0.908d |
| Yes or consumed | 78 (48.2) | 16 (47.1) | |
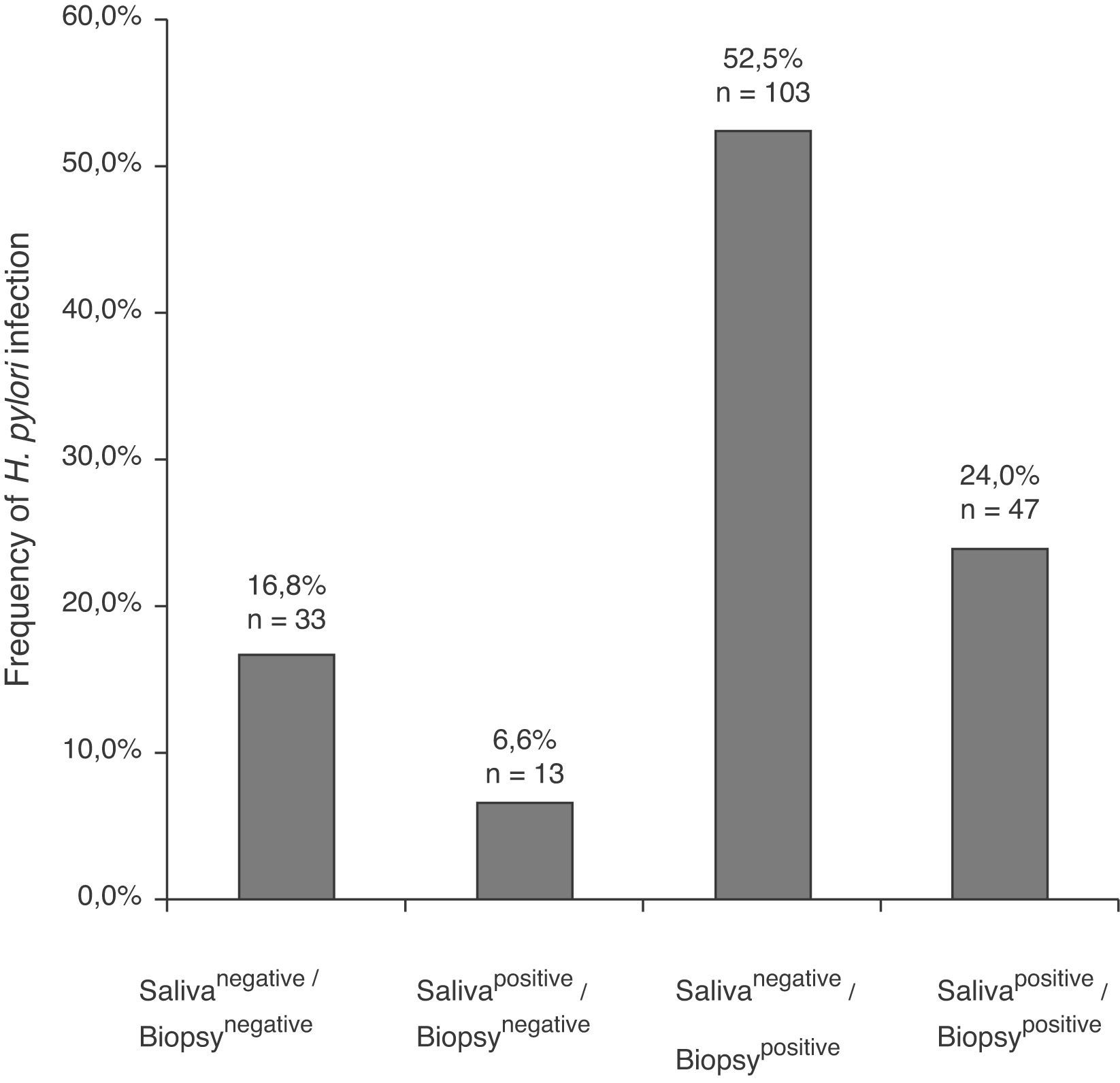
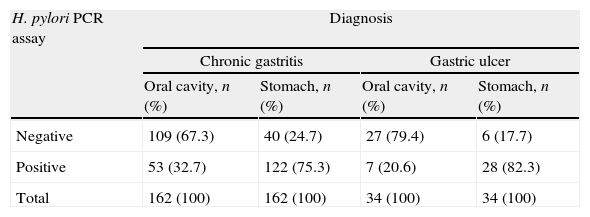
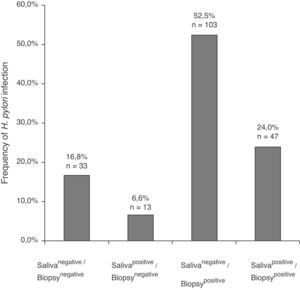
In the 196 patients, H. pylori prevalence was 76.5% in the stomach and 30.6% in saliva. The H. pylori DNA in saliva samples was detected in 32.7% (53/162) of chronic gastritis patients and in 20% (7/34) of gastric ulcer patients (Table 2); 78.3% (47/60) of patients with saliva H. pylori-positive had gastric infection (kappa=0.018), (Fig. 1). The primers used only amplified the rRNA 16S gene of H. pylori.
Frequency of H. pylori in saliva and gastric biopsies of patients with chronic gastritis and gastric ulcer.
| H. pylori PCR assay | Diagnosis | |||
| Chronic gastritis | Gastric ulcer | |||
| Oral cavity, n (%) | Stomach, n (%) | Oral cavity, n (%) | Stomach, n (%) | |
| Negative | 109 (67.3) | 40 (24.7) | 27 (79.4) | 6 (17.7) |
| Positive | 53 (32.7) | 122 (75.3) | 7 (20.6) | 28 (82.3) |
| Total | 162 (100) | 162 (100) | 34 (100) | 34 (100) |
The vacA allelic variants were determined in 78.3% (47/60) of patients with saliva H. pylori-positive, Fig. 2. The s1m1 and s1m2 genotypes were detected alone or coexisting in the saliva of 41.5% (22/53) of patients with chronic gastritis and 47.2% (25/53) had more than one vacA genotype; 36% (9/25) had s1m1/s1m2 and 16% (4/25) had s1m1/s2m1. In saliva H. pylori-positive of patients with gastric ulcer vacA s1m1 was found in 28.6% (2/7) of samples and vacA s1m2 was found in 14.3% (1/7). One patient with H. pylori vacA s1m1/s1m2 genotypes in saliva was also found to have s1m1 in stomach (Table 3).
Fragments of s and m alleles from H. pylori vacA generated by nested PCR. MWM 1-kilobase DNA ladder; C− negative control (contain all the necessary components except template DNA); C+ positive control Helicobacter pylori J99, (HPJ99) genotype s1m1. Genomic DNA extracted from saliva or gastric biopsies.
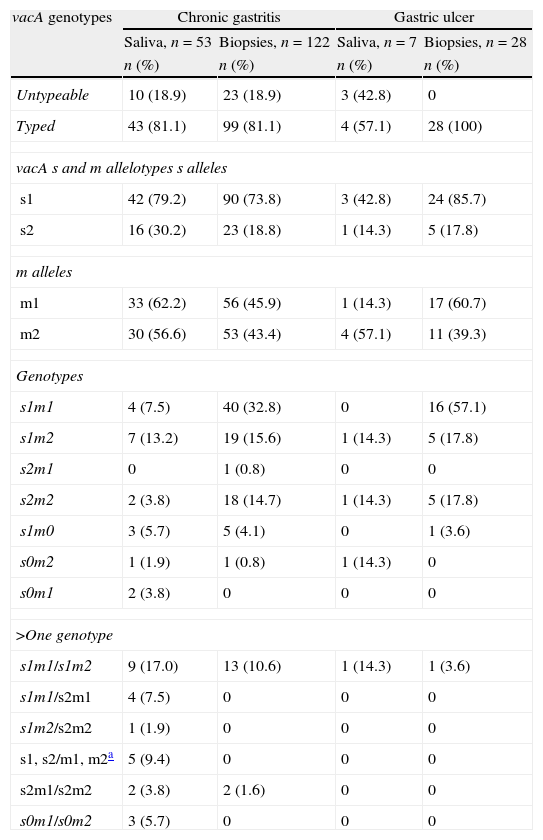
Frequency of vacA genotypes in patients with chronic gastritis and gastric ulcer positive for H. pylori DNA in saliva and stomach.
| vacA genotypes | Chronic gastritis | Gastric ulcer | ||
| Saliva, n=53 | Biopsies, n=122 | Saliva, n=7 | Biopsies, n=28 | |
| n (%) | n (%) | n (%) | n (%) | |
| Untypeable | 10 (18.9) | 23 (18.9) | 3 (42.8) | 0 |
| Typed | 43 (81.1) | 99 (81.1) | 4 (57.1) | 28 (100) |
| vacA s and m allelotypes s alleles | ||||
| s1 | 42 (79.2) | 90 (73.8) | 3 (42.8) | 24 (85.7) |
| s2 | 16 (30.2) | 23 (18.8) | 1 (14.3) | 5 (17.8) |
| m alleles | ||||
| m1 | 33 (62.2) | 56 (45.9) | 1 (14.3) | 17 (60.7) |
| m2 | 30 (56.6) | 53 (43.4) | 4 (57.1) | 11 (39.3) |
| Genotypes | ||||
| s1m1 | 4 (7.5) | 40 (32.8) | 0 | 16 (57.1) |
| s1m2 | 7 (13.2) | 19 (15.6) | 1 (14.3) | 5 (17.8) |
| s2m1 | 0 | 1 (0.8) | 0 | 0 |
| s2m2 | 2 (3.8) | 18 (14.7) | 1 (14.3) | 5 (17.8) |
| s1m0 | 3 (5.7) | 5 (4.1) | 0 | 1 (3.6) |
| s0m2 | 1 (1.9) | 1 (0.8) | 1 (14.3) | 0 |
| s0m1 | 2 (3.8) | 0 | 0 | 0 |
| >One genotype | ||||
| s1m1/s1m2 | 9 (17.0) | 13 (10.6) | 1 (14.3) | 1 (3.6) |
| s1m1/s2m1 | 4 (7.5) | 0 | 0 | 0 |
| s1m2/s2m2 | 1 (1.9) | 0 | 0 | 0 |
| s1, s2/m1, m2a | 5 (9.4) | 0 | 0 | 0 |
| s2m1/s2m2 | 2 (3.8) | 2 (1.6) | 0 | 0 |
| s0m1/s0m2 | 3 (5.7) | 0 | 0 | 0 |
s0: non-typeable for signal region.
m0: non-typeable for middle region.
In the stomach of patients with chronic gastritis, H. pylori vacA s1m1 was detected in 43.4% (53/122) and vacA s1m2 was found in 26.2% (32/122). In 15 of the 122 biopsies H. pylori-positive two genotypes were found and 86% (13/15) of them were vacA s1m1/s1m2. In patients with ulcer, 60.7% (17/28) of gastric infections were due to H. pylori vacA s1m1 and 21.4% (6/28) to s1m2 (Table 3). There were eight strains untypeable for the s region and nine strains untypeable for m region. It was not possible to identify vacA genotype in eight biopsies of salivapositive/biopsypositive patients but it was done in 7 of 8 saliva samples. There were significant differences among vacA genotypes found in saliva and gastric biopsies in patients with chronic gastritis (p=0.001 by Fisher test) and gastric ulcer (p=0.002 by Fisher test).
There was 51.1% (24/47) agreement between genotypes found in biopsy and their corresponding saliva DNA sample in H. pylori-positive patients (kappa=0.095). Two genotypes in saliva and one or both in the stomach were detected simultaneously in 27.6% (13/47) of the patients.
DiscussionThe five central findings in this study were (1) that in 24% of the patients H. pylori DNA was simultaneously found in saliva and gastric biopsy, (2) 6% of the patients were H. pylori DNA salivapositive/biopsynegative, (3) the presence of >1 vacA genotype is commonly found in saliva and biopsy of the same patient, (4) both s1m1 and s1m2 were the most frequent genotypes in both groups of patients with preponderance of s1m1 in saliva and in gastric biopsy, (5) 51.1% of the salivapositive/biopsypositive patients presented the same genotypes in both sites. Our first and second findings suggest that gastric infection is not the only responsible one for the occurrence of H. pylori in the oral cavity and it is possible that the bacterium might reach the mouth in various ways besides gastric reflux. These findings are in agreement with the low correlation between gastric infection and the occurrence of H. pylori in the mouth, as it has previously been reported.6,7,22,23 Gastric lesions may be caused by factors that predispose to ulceration of the gastric mucosa such as stomach acid, stress, alcohol, tobacco, and non-steroidal anti-inflammatory drugs (NSAIDs), Table 3. Some of these factors were not investigated in this work. We may not rule out the possible participation of H. pylori in the start of the diseases and we cannot exclude bias of information about treatment by the patients.
González-Valencia et al.24 reported that gastric infection with more than one vacA genotype of H. pylori in the same biopsy is more frequent in Mexico than in other countries and our findings support that observation. The results of the present study show that a significant proportion of saliva samples harbored >1 vacA genotype of H. pylori which were found simultaneously in biopsy of the same patient. Okuda et al.25 found the same H. pylori strain in both the oral cavity and stomach and showed that the bacterium is trapped in biofilms or macroaggregates with other buccal bacteria and that H. pylori adheres to saliva MUC5B mucin.9,22,25 It is likely that different H. pylori strains reach the oral cavity by different routes, person to person, vomiting or gastroesophageal reflux, and it is also possible that they stay in saliva and dental plaque long enough to reach the stomach. Our results sustain the hypothesis that oral colonization can be a reservoir of H. pylori and source of stomach infection or re-infection.2,7,23
This research confirms that vacA s1m1 and s1m2 are the most frequent genotypes found in the gastric infections of Mexican patients with gastroduodenal pathology.10,26,28,29vacA s1m1 was found associated with a more severe form of gastric pathology in other studies.10,15,16,26
Fernández-Tilapa et al.2 found s1 and m1 alleles in the oral cavity of asymptomatic patients and in this study their combinations were found in the mouth and in the stomach of patients with chronic gastritis and gastric ulcer. To reach the stomach, H. pylori must pass through the mouth and it is likely that interpersonal relationships contribute to person to person transmission.
There were 17 strains untypeable for s or m region. Similar findings have been described previously in Mexican populations, and the possible existence of different subfamilies of vacA26,27 had been suggested. Furthermore, H. pylori has at least two copies of 16S and 23S rRNA genes but only one vacA gene.30 Probably, the copies number of vacA gene in the samples could not be detected by nested PCR. On the other hand, it has revealed the existence of indistinguishable genotypes from oral cavity and gastric mucosa samples of the same patient.8 Interestingly, we were able to identify vacA genotypes in saliva DNA but it was not possible to do it in corresponding biopsies H. pylori-positives. These findings suggest that search and genotyping of H. pylori must be simultaneous in mouth and stomach.
H. pylori prevalence in saliva and in gastric biopsy was 30.6% and 75%, respectively, and higher than reported by Morales-Espinosa et al.29 in Mexican patients with gastroesophagic disease as well as that reported by other researchers in various countries.8,25 Amplification of H. pylori DNA in oral samples by PCR techniques, that have been used to circumvent the culturing problems, has demonstrated highly variable detection rates ranging from 0% to 100%.6–8,11,23 These discrepancies might be explained by differences such as population origin, number and diagnosis of the patients, methodological differences regarding the collection and processing of samples, oral hygiene habits or fasting condition of the patients at the time of sampling, and specificity of the PCR primers. The detection of H. pylori may also be influenced by variations in the number of bacteria in saliva considering the flow and replacement rates.
The geographic distribution of gastric infection by H. pylori depends from the interactions between the bacterium and the host as well as the environment.
In the present study, for detection of H. pylori and the molecular characterization of vacA PCR and nested PCR were used, respectively. The PCR assay has been demonstrated to be more efficient for the detection of H. pylori in clinical specimens, compared to the rapid urease test in dental plaque and gastric biopsies, as well as to the histopathological examination of the gastric mucosa.2,7,8,10,23
The presence of similar vacA genotypes in the mouth and in gastric biopsy of the same patient and oral bacterial colonization in patients without gastric infection suggests a role of the oral cavity in infection and re-infection. In our knowledge, this is the first study that reports the correlation between vacA genotypes in saliva and gastric biopsy of patients with chronic gastritis and gastric ulcer.
H. pylori infection, alcohol, soda and coffee consumption are risk factors for peptic acid disorders. Nevertheless, we did not find a correlation of these beverages with diagnosis even though they might change the mouth pH and, thus, oral colonization by H. pylori.
ConclusionWe conclude that vacA s1m1 and s1m2 genotypes alone or in coexistence are most prevalent in saliva and in gastric biopsy of patients with chronic gastritis and gastric ulcer and they are simultaneously found in saliva and in gastric biopsy of the same patient. Even though H. pylori is found mainly in the mouth of patients with disease and gastric infection, there is a high number of patients negative for the bacteria in the stomach that harbor H. pylori in the mouth. This fact might be an obstacle for bacterial eradication and at the same time a risk for gastrointestinal re-infection after therapy.
Our findings indicate that gastric reflux is not the only route by which H. pylori reaches the mouth and that detection and genotyping in mouth and in stomach are complementary tests.
Conflict of interestThe authors declare no conflict of interest in the publication of this research.
We are grateful to Dr. Reyes Betancourt Linares, as well as the nurses and support personnel of the Specialized Unit of Gastroenterology Endoscopy from Chilpancingo, who assisted in obtaining samples. We also want to thank Martín O. Morrugares-Ixtepan, Specialist in Pathological Anatomy with subspecialty in Oncological Pathology, who was responsible for the histopathological diagnoses. The excellent technical assistance of QBP. Josefina Atrisco Morales is greatly appreciated. We thank especially Jon Gonzalez Santamaria, for checking grammar of this paper.
This work was supported by the Secretaría de Educación Pública, via PIFI 2009 and by the Programa de Apoyo a la Reincorporación de Exbecados PROMEP 2009, key PROMEP UAGUER-EXB-096. During our study Adolfo Román-Román was a grant recipient from the Universidad Autónoma de Guerrero, México, and Silvia Giono-Cerezo was a grant recipient from the CONACyT, EDI, Cofaa, México.