The AMR Direct Flow Chip assay allows the simultaneous detection of a large variety of antibiotic resistance genetic markers. To assess this kit's performance, we use isolated colonies as starting material. The assay has been approved by the European Economic Area as a suitable device for in vitro diagnosis (CE IVD) using clinical specimens.
MethodsA total of 210 bacterial isolates harbouring either one or more antimicrobial resistance genes including plasmid-encoded extended-spectrum β-lactamases (SHV, CTX-M) and carbapenemases (GES, SME, KPC, NMC/IMI, SIM, GIM, SPM, NDM, VIM, IMP, and OXA), mecA, vanA and vanB, and 30 controls were included.
ResultsThe assay displayed a sensitivity and specificity of 100% for all target genes included in the array.
ConclusionThe AMR Direct Flow Chip Kit is an accurate assay for detecting genes which commonly confer resistance to β-lactams and vancomycin from isolated colonies in culture of Gram-positive and Gram-negative bacteria.
El ensayo «AMR Direct Flow Chip Kit» permite detectar simultáneamente la presencia de una gran variedad de marcadores genotípicos de resistencia bacteriana. Evaluamos su rendimiento utilizando colonias aisladas como material de partida. El ensayo aludido ha sido aprobado por el Área Económica Europea como un dispositivo adecuado para el diagnóstico in vitro (CE IVD) utilizando muestras clínicas.
MétodosEl estudio ha incluido 210 aislados bacterianos con uno o más genes de resistencia a los antimicrobianos, incluidos genes plasmídicos que codifican β-lactamasas de espectro extendido (SHV y CTX-M) y carbapenemasas (GES, SME, KPC, NMC/IMI, SIM, GIM, SPM, NDM, VIM, IMP y OXA), mecA, vanA y vanB, y 30 controles.
ResultadosEl ensayo mostró una sensibilidad y especificidad del 100% para todos los genes diana incluidos en la matriz.
ConclusiónEl «AMR Direct Flow Chip Kit» es un ensayo fiable para la detección de genes que comúnmente confieren resistencia a β-lactámicos y vancomicina en bacterias grampositivas y gramnegativas a partir de colonias aisladas en cultivo.
Timely detection and characterization of antimicrobial resistance is crucial for both adequate management of bacterial infections and epidemiological control of multi-resistant bacteria spread.1 Molecular methods allow rapid and sensitive detection of genetic elements linked with antimicrobial resistance traits. Among these, multiplex PCR combined with DNA microarray chips has emerged as a robust technology to simultaneously screen for the presence of a vast number of target genes in a single test, using either clonal culture material or clinical specimens.2 In recent years, a number of both in-house and commercially-available DNA microarray assays have been developed and evaluated for detection of carbapenemase and extended-spectrum β-lactamases (ESBL) genes in Enterobacteriaceae, Pseudomonas aeruginosa and Acinetobacter baumannii, and methicillin-resistance in Staphylococcus aureus.3–12
Here, we evaluated the performance of the Antimicrobial Resistance (AMR) Direct Flow Chip (Máster Diagnóstica, Granada, Spain), a DNA microarray-based assay for antimicrobial resistance gene detection from bacterial isolated colonies. This assay has been approved by the European Economic Area as a suitable device for in vitro diagnosis (CE IVD) using clinical specimens (rectal and nasopharyngeal swabs, blood cultures flagged as positive), but not isolated colonies as starting material.
MethodsThe evaluation panel consisted of 240 archived clinical isolates of which 160 were isolated at the Microbiology Service of Hospital Clínico Universitario and 50 at the Microbiology Service of Consorcio Hospital General Universitario. The remaining 30 isolates were kindly provided by Dr. Antonio Galiana (Department of Microbiology, Hospital General Universitario de Elche) and Professor Álvaro Pascual (Unidad de Enfermedades Infecciosas, Microbiología y Medicina Preventiva, Hospital Universitario Virgen Macarena, Sevilla, Spain). The panel included 210 isolates (Table 1) possessing one or more resistance genes potentially detectable by the AMR assay, namely carbapenemase or ESBL-producing Enterobacteriaceae (n=104), P. aeruginosa (n=50), and A. baumannii (n=13), methicillin-resistant S. aureus-MRSA- (n=7), vancomycin-resistant Enterococcus faecalis (n=3) or faecium (n=33) – VRE.
Antimicrobial resistance genes profiles of bacterial species included in the study.
| Bacterial species (no. of isolates) | Resistance genes detected (no. of isolates) |
|---|---|
| Gram-negative | |
| Klebsiella pneumoniae (65) | SHV, CTX-M, OXA-48 (21) |
| SHV, CTX-M (13) | |
| CTX-M, OXA-48 (8) | |
| SHV (6) | |
| SHV, VIM (4) | |
| SHV, CTX-M, NDM (3) | |
| SHV, OXA-48 (2) | |
| SHV, KPC (2) | |
| NMC/IMI (2) | |
| VIM (2) | |
| CTX-M, NDM, OXA-48 (1) | |
| KPC (1) | |
| Pseudomonas aeruginosa (50) | VIM (43) |
| IMP (3) | |
| SPM (2) | |
| GIM (1) | |
| VIM, OXA-58 (1) | |
| Escherichia coli (14) | CTX-M (6) |
| KPC, OXA-58 (1) | |
| KPC, NDM (1) | |
| SHV, GES, OXA-48, OXA-51 (1) | |
| SME (1) | |
| VIM (1) | |
| NDM (1) | |
| OXA-48 (1) | |
| CTX-M, OXA-48 (1) | |
| Enterobacter cloacae (13) | CTX-M (3) |
| NMC/IMI (2) | |
| KPC (2) | |
| VIM (1) | |
| CTX-M, VIM (1) | |
| SHV, GES, OXA-48 (1) | |
| CTX-M, GES (1) | |
| SHV, OXA-51 (1) | |
| OXA-48 (1) | |
| Acinetobacter baumannii (13) | OXA-23, OXA-51 (6) |
| OXA-24, OXA-51 (2) | |
| OXA-58, OXA-51 (1) | |
| SIM, OXA-51 (1) | |
| SIM (1) | |
| OXA-51 (1) | |
| OXA-23 (1) | |
| Serratia marcescens (4) | CTX-M, OXA-48 (3) |
| SME (1) | |
| Enterobacter asburiae (3) | NMC/IMI (2) |
| KPC (1) | |
| Citrobacter freundi (2) | NDM (1) |
| SHV, CTX-M, VIM (1) | |
| Citrobacter amalonaticus (1) | VIM, OXA-48 (1) |
| Morganella morganii (1) | OXA-48 (1) |
| Klebsiella variicola (1) | SHV (1) |
| Gram-positive | |
| Enterococcus faecium (33) | vanA (33) |
| Enterococcus faecalis (3) | vanA (1) |
| vanB (2) | |
| Staphylococcus aureus (7) | mecA (7) |
We included as negative controls 30 isolates displaying either a wild-type resistance phenotype, including Escherichia coli (n=5), Proteus vulgaris (n=1), Klebsiella oxytoca (n=1), Enterobacter cloacae (n=1), Enterobacter aerogenes (n=1), Serratia marcescens (n=1), MSSA (n=3), vancomycin-susceptible E. faecium (n=3), or a resistant phenotype mediated by mechanisms undetectable by the assay, namely carbapenem-resistant P. aeruginosa (n=8) with a negative Rapidec Carba NP test (Biomerieux, L’Etoile, France), β-lactam resistant Enterobacteriaceae mediated by plasmidic AmpC (K. pneumoniae, n=1), or derepressed chromosomal AmpC (E. cloacae, n=4) and E. casseliflavus (vanC, n=1). In addition, the ATCC KPC-producer K. pneumoniae reference strain (BAA-1705) and the non-KPC producer carbapenem-resistant K. pneumoniae strain (BAA-1706) were also included.
Bacterial isolates resistance mechanisms were previously determined by means of a variety of phenotypic and genotypic methods, including among the latter the Eazyplex® SuperBug CRE (Amplex, Giessen, Germany), targeted real-time PCR assays,13,14 and PCR coupled to sequencing.13
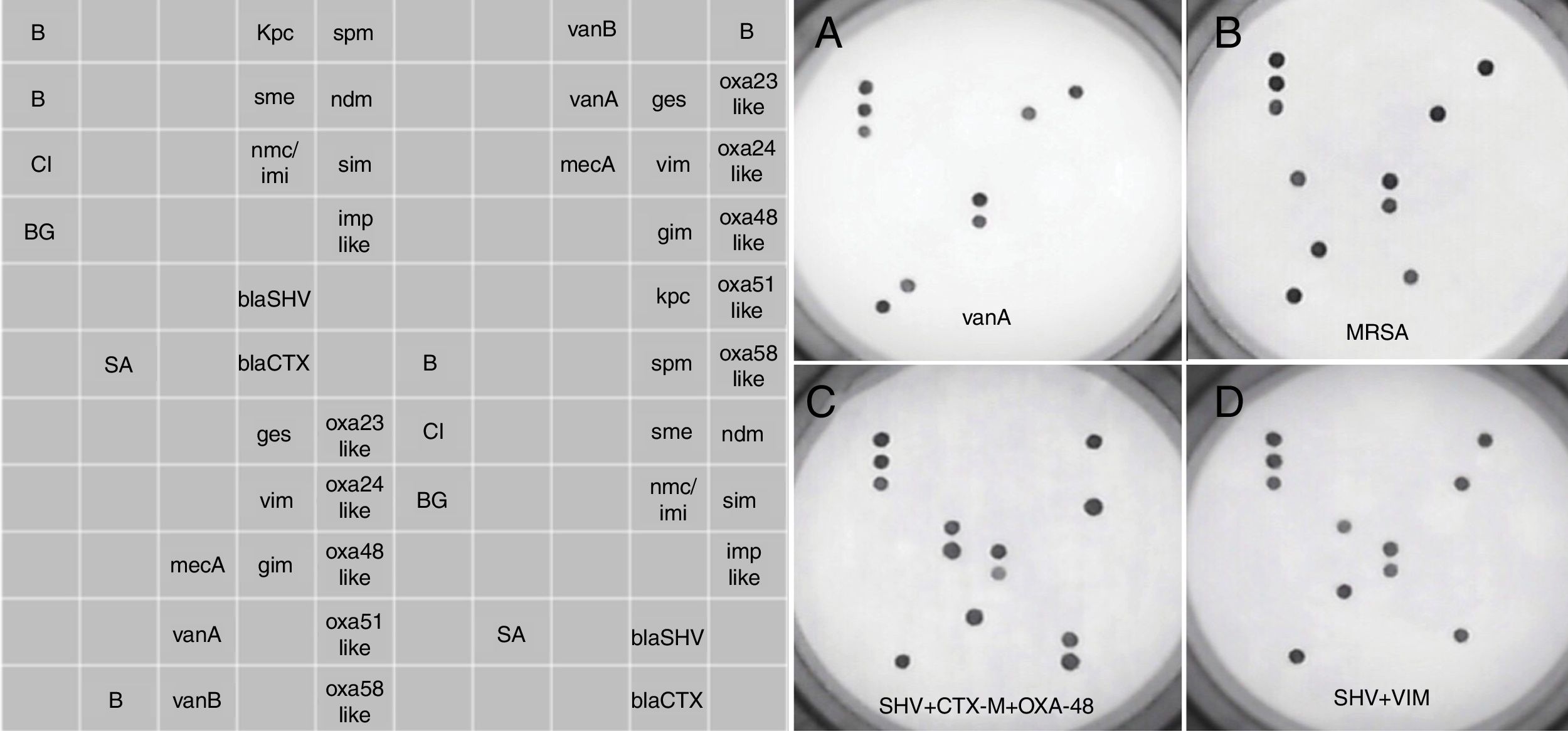
Retrieved isolates were cultured overnight onto 5% Columbia blood agar. A single colony was homogenized in 50μl of sterile distilled water, of which a volume of 5μl was directly used for AMR testing, without prior DNA extraction. The assay was performed according to the manufacturer's instructions, previously detailed in Galiana et al.15 for the Sepsis Flow Chip (Máster Diagnóstica, Granada, Spain), which has the same processing routine as the AMR assay. The AMR assay is based on a multiplex PCR amplification using biotinylated primers followed by an automatic reverse hybridization in membrane containing specific probes to detect 20 known resistance genes in isolated Gram-negative bacilli and Gram-positive cocci. Specifically, the assay is able to detect class A carbapenemases including GES (allelic variants 1–26), SME (1–5), KPC (1–23), NMC/IMI (1–9), class B carbapenemases including SIM, GIM (allelic variants 1 and 2), SPM, NDM (1 to 16), VIM (1–46), IMP (allelic variants 1, 2, 3, 5, 6, 8, 9, 10, 11, 15, 19, 20, 21, 24, 25, 28, 29, 30, 40, 41, 42 and 47), class D carbapenemases including OXA23-like (14 allelic variants), OXA24-like (7 allelic variants), OXA48-like (4 allelic variants), OXA51-like (67 allelic variants), OXA58-like (4 allelic variants), ESBLs blaSHV (allelic variants detected are not disclosed by the manufacturer), ESBL blaCTX-M (allelic variants detected are not disclosed by the manufacturer), S. aureus mecA, and vanA and vanB genes. The assays do not distinguish between β-lactamases allelic variants. Positive signals are visualized through a colorimetric immunoenzymatic reaction in a chip membrane by the HS24 hybridization platform, which includes a built-in camera that captures the image of the chip and analyzes the dot pattern by means of the hybrisoft software. See image examples in Figure 1. The assay can be completed in 4h.
Prototypical DNA microarray pictures obtained with the AMR Direct Flow Chip assay. The left panel shows the distribution of complementary oligonucleotides targeting specific antibiotic resistance genes sequences. The right panels (A–D) show the results for different bacterial species harbouring one or more antimicrobial resistance genes. B, hybridation control; CI, exogenous amplification control; BG, endogenous amplification control; SA, Staphylococcus aureus; MRSA, methicillin-resistant Staphylococcus aureus.
With regard to Gram-positive bacteria, the AMR microarray correctly identified the presence of the mecA, vanA and vanB genes in all isolates (MRSA, n=7 and VRE, n=36, respectively). The control isolates, including one E. casseliflavus (vanC) isolate, tested negative.
As for Gram-negative bacteria, the assay detected all carbapenemase genes carried by P. aeruginosa, A. baumannii and Enterobacteriaceae isolates in our panel, including GES (n=3), SME (n=2), KPC (n=8), NMC/IMI (n=6), SIM (n=2), GIM (n=1), SPM (n=2), NDM (n=7), VIM (n=55), IMP (n=3) and OXA (OXA-23, OXA-24, OXA-48, OXA-51, OXA-58; n=67), present individually or in combination (Table 1). Likewise, results obtained with the AMR assay for ESBLs (CTX-M; n=62 and SHV-ESBL; n=56) in Enterobacteriaceae were fully concordant with those expected, irrespective of whether these genes were harboured in isolation, or in combination (Table 1). None of the control isolates tested positive in the assay.
DiscussionThe AMR Direct Flow Chip has been approved by the European Economic Area as a suitable device for in vitro diagnosis (CE IVD) using clinical specimens, but not isolated colonies as starting material. Although it can be presumed that the performance of the assay with isolated colonies should be at least as good as with clinical samples, validation for indications outside the technical data sheet is warranted.
In our experience, the AMR Direct Flow Chip Kit demonstrates 100% sensitivity and specificity for detection of locally-relevant antimicrobial resistance genes from Gram-positive and Gram-negative bacteria using isolated colonies. Nevertheless, it must be highlighted the fact that the kit has been designed and validated to detect all variants of SHV, the original SHV-1 and all mutated forms of the gene, without distinction between them. For this reason, detection of SHV by AMR Direct Flow Chip Kit would not necessarily indicate a phenotypic evidence of extended-spectrum β-lactamase production.
In this sense, the AMR assay performs comparably, or even better, than a widely-used series of commercially-available DNA arrays, the Check-MDR array versions CT102, CT103 and CT103XL (Check Point, Wageningen, The Netherlands) designed to detect common and clinically relevant carbapenemase and ESBLs, whose reported sensitivity and specificity values vary between 85% and 100% for both parameters depending upon the targeted resistance gene (lowest values for KPC carbapenemases).3–7,10,12 A non-negligible advantage of the AMR Direct Flow Chip over the aforementioned microarrays is that it skips the DNA extraction step. In contrast, a noteworthy disadvantage of this assay is its inability to detect certain antibiotic resistance genes widely prevalent among Enterobacteriaceae species in our setting (i.e. TEM-derived ESBL and plasmidic AmpCs).
The current study has several limitations that deserve comment. First, the distribution of bacterial species in our panel was rather unbalanced with a clear predominance of K. pneumoniae and P. aeruginosa isolates; second, bacterial species harbouring certain resistance genes (i.e. NDM, KPC and vanB) were scarcely represented; third, the isolates in our collection were not subjected to clonality analysis. Although care was taken to include isolates recovered from two different hospitals over a relatively long time span, and displaying a wide variety of antimicrobial resistance phenotypic patterns (not shown) clonality bias cannot be ruled out. All the aforementioned limitations may undermine the robustness of our conclusions. In summary, our data indicate that the AMR Direct Flow Chip Kit is an accurate assay for detecting widely-spread genes conferring resistance to β-lactams and vancomycin from isolated colonies of Gram-positive and Gram-negative bacteria.
FundingThis research did not receive any specific grants from funding agencies in the public, commercial, or not-for-profit sectors.
Conflict of interestThe authors report no conflicts of interest.
We thank Juan Miguel Moya, Carme Salvador and Javier Colomina for technical assistance and Dr. Antonio Galiana and Professor Álvaro Pascual for kindly providing bacterial isolates with defined antibiotic resistance genes. We also thank Vitro S.A. (Madrid, Spain) for the kind donation of the reagents used in this study.