The Umbilical Cord Blood Units (UCBU) for transplant are a therapeutic possibility for patients with a wide range of onco-hematologic disorders, especially in children. In Mexico, 48.5% of oncological diseases in children from 1 to 4 years old are leukemias; while in patients from 5 to 14 and 15 to 24 years of age lymphomas and leukemias are predominant and represent the second and third causes of death in these age groups, respectively. Therefore, is it necessary to have registries of UCBU to ensure the representation of the genetic diversity in Mexico in order to attend this requirement.
ObjectiveTo estimate the genetic diversity of HLA Class I (A, B) and Class II (DRB1) loci in cryopreserved UCBU of the Cord Blood Bank (CBB) at the National Center of Blood Transfusion (NCBT).
MethodsHLA typing of 533 UCBU for transplant was performed at the Research Department (evaluated by “Los Angeles Ca. Inmunogenetics Center”). Class I HLA-A, HLA-B and Class II HLA-DRB1 typing was performed using medium resolution Sequence-Specific Primer (SSP). In cases of an ambiguity by SSP; Sequence-Specific Oligonucleotide (SSO) method was carried out.
Results46.5% of the UCBU were obtained from Mexico City donors, 30.95% from the State of Mexico, 8.06% Puebla, 6.37% Morelos and 3.37% from Veracruz. The remaining UCBU 4.75% were represented by other states of the country. The most frequent loci for the HLA-A founded were *02/24, *02/68, *02/02, *02/30, *01/02, *02/31; for HLA-B, *35/39, *15/35, *35/40, *39/44, *07/35, *35/48, *39/40 and for HLA-DRB1, *04/08, *04/07, *04/15, *04/15, *04/03, *04/14. The genetic distances analysis showed that the top five populations analyzed in this study are significatively different from each other.
ConclusionsThe majority of the genotypes found suggest Amerindian and European origins and in a lesser proportion Oriental and African. The NCBT is therefore establishing agreements with different states of Mexico to promote the donation of UCBU in order to enrich the genetic diversity in the archives of the NCBT.
Las Unidades de Sangre de Cordón Umbilical (USCU) para trasplante constituyen una posibilidad terapéutica para pacientes con trastornos onco-hematológicos, especialmente en niños. En México 48.5% de las enfermedades oncológicas en niños de 1 a 4 años son las leucemias; mientras que en pacientes de 5-14 y 15-24 años de edad, los linfomas y leucemias son predominantes y representan la segunda y tercera causa de muerte en estos grupos de edad, respectivamente. Por lo anterior, es necesario contar con registros de UCBU para asegurar la diversidad genética de estas unidades en el país.
ObjetivoEstimar la diversidad genética del HLA de Clase I (A, B) y Clase II (DRB1) en USCU criopreservadas en el Banco de Sangre de Cordón (BSC) del Centro Nacional de Transfusión Sanguínea (CNTS).
MétodoLa tipificación del HLA de 533 UCBU se llevó a cabo en el Departamento de Investigación (evaluado por “El Centro de Inmunogenética de Los Angeles Ca.”). La tipificación del HLA de Clase I y II se realizó mediante el método de mediana resolución “Secuencia-Cebador-Específico” (SSP). En los casos de ambigüedades por SSP; se empleó el método de Oligonucleótido-Secuencia-Específico (SSO).
ResultadosEl 46.5% de las UCBU fueron colectadas en la Ciudad de México, 30.95% en el Estado de México, 8.06% Puebla, 6.37% Morelos y 3.37% de Veracruz. El resto de las UCBU (4.75%) están representadas por otros estados del país. El loci más frecuente encontrado para HLA-A fue *02/24, *02/68, *02/02, *02/30, *01/02, *02/31; para HLA-B, *35/39, *15/35, *35/40, *39/44, *07/35, *35/48, *39/40 y para el HLA-DRB1 fueron *04/08, *04/07, *04/15, *04/15, *04/03, *04/14. El análisis de las distancias genéticas mostró que las cinco poblaciones analizadas son significativamente diferentes unas de otras
ConclusionesLa mayoría de los genotipos encontrados sugieren orígenes Amerindios y Europeos y en menor proporción Orientales y Africanos. Por lo anterior, el CNTS está estableciendo convenios con los diferentes estados de la República Mexicana para promover la donación de USCU con el fin de enriquecer la diversidad genética en sus archivos.
Genetic characterization of the HLA system is needed in the selection study of donors/receptors in hematopoietic transplants. The Human Leukocyte Antigens (HLA) include a family of genes located in the short arm of chromosome 6 and represent one of the most polymorphic systems of the human genome.1 For patients with diseases of onco-hematological origin, there are three possible sources of progenitor cell donors: bone marrow or mobilized blood from a member of the family (parents or siblings), unrelated adult donors and cryopreserved Umbilical Cord Blood Unit (UCBU) of a Cord Blood Bank (CBB).2 A low rate of development of Graft Versus Host Disease (GVHD) is a major advantage of the transplantation of UCBU, whereas the delayed engraftment due to the limited dose of cells remains as the major drawback.3 The genetic pool in urban areas of Mexico such as Mexico City, Guadalajara and Monterrey has been estimated from genetic studies on blood groups, serum proteins, and DNA markers.4–7 The genetic admixture of the Mexican population is complex, as well as its demographic history, as shown by some studies on mitochondrial DNA (mtDNA).8,9 Various estimates show an unequal distribution in HLA genotypes throughout the national territory, with a tendency toward a greater African component to the southeastern coast and parts of the Midwest and a higher proportion of genotypes from Europe to the north of the territory.10,11 The unequal access to basic services has caused rural migration to the city of Mexico and to other rural zones. This has changed the genetic distribution throughout the country.12,13 For all these reasons it is necessary to have CBB with a reservoir of UCBU with genetic representation at national level, to ensure the availability of UCBU for transplant. The aim of this study is to describe the frequency of HLA genotypes obtained by the analysis of the distribution of the class I (A and B) and class II (DRB1) genotypes in the cryopreserved UCBU in the Umbilical Cord Blood Bank (UCBB) of the NCBT from different geographical regions (Mexico City, State of Mexico, Puebla, Morelos, Veracruz, Zacatecas, Hidalgo, Querétaro, Tlaxcala, Oaxaca, Chihuahua, Colima, Guanajuato, Guerrero and Yucatán) in order to estimate the genetic diversity.
Materials and methodsEthical aspectsThe Research Ethics Committee of the NCBT approved this protocol. Additionally, the collection and presentation of data were carried out under the observance of the principles of confidentiality and discretion outlined by the Federal Law of Accountability and Access to Public Government Information.
Umbilical Cord Blood Units sample533 Umbilical Cord Blood (UCB) samples (1.5mL) collected in a period of 12 years (2003–2014) send to the NCBT from various states of Mexico (unrelated individuals), were collected in sterile cryotubes in CPD (Citrate-Phosphate-Dextrose) and sent to the Histocompatibility Laboratory of the NCBT to type the HLA-A, HLA-B and HLA-DRB1 genotypes at medium resolution. The size of sample of UCBU analyzed in this work, were those with the necessary requirements (CD34+ cells, white blood cells, clonality, infectious serology and microbiology negative) for used for transplantation purposes.
DNA extractionAliquots of 200μL of UCB were used for the extraction of total DNA, using the DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) following the instructions provided by the manufacturers. DNA purity was determined by 180/160nm proportion (≈1.8) before amplification.
Class I and II HLA typingThe HLA Class I and Class II typing was performed by PCR-SSP (Polymerase Chain Reaction-Sequence Specific Primer) (Micro SSP-One Lambda Inc., Canoga Park, CA) and Sequence-Specific Oligonucleotide (SSO) methods (Labtype SSO kit, One Lambda, Inc., Canoga Park, CA). In the SSP method, the plate containing amplicons was separated and analyzed in horizontal electrophoresis in 2.5% agarose gels at 150 volts for 4min using 1×Tris–Borate–EDTA buffer (TBE). In the case of the SSO method, total DNA of UCB was amplified in a master mix (D-mix and primers for the A, B and DRB1 loci) plus 1 U of Taq DNA polymerase (Thermo Fisher Scientific Inc. USA) in a final volume of 20mL. The mixtures of PCR were amplified of exons 2 and 3 of the A and B and exon 2 of the DRB1 loci as recommended by the manufacturer using a Touchgene Gradient thermal cycler Gene Amp® PCR System 9700 (Applied Biosystems). The amplified exons were hybridized using probe conjugated beads, which have the same nucleotide sequence as the amplified exon (A, B and DRB1). The amplicons were used for hybridization with different oligonucleotide-conjugated beads which bind to specific complementary sequences. This reaction is revealed by the binding of biotinylated probes that react with streptavidin-phycoerythrin (SAPE), which in turn binds to fluorescently-labeled beads of different colors. Data were analyzed using the HLA Fusion software-Labtype (One Lambda® – San Diego, CA, USA).
Statistical analysisThe loci (HLA-A and HLA-B) and locus (HLA-DRB1) frequencies were calculated by a direct counting method and all data were entered into computer using a spreadsheet program (Excel, Microsoft Corp., Redmond, WA). Statistics data were calculated to describe the locus frequencies in the cryopreserved UCBU. These calculations included the median frequencies and the most frequent haplotypes. A Hardy–Weingberg Test was performed between the populations with n≥18 individues for each loci studied. The linkage disequilibrium (LD) between pairs of loci was calculated using the genetic package Genepop (http://genepop.curtin.edu.au).
Populations comparisonsUsing loci frequencies of HLA-A, HLA-B and HLA-DRB1 locus the significant difference in the different states of the Mexican Republic was performed by analysis of variance (ANOVA) and based on the least significant difference using linear model,14 with R Stats Package v. 3.0.2.15
Genetic distances measurementThe HLA genotype frequencies data were used to estimate Cavalli-Sforza chord distance,16 between the top five populations analyzed in this study (Mexico City, State of Mexico, States of: Puebla, Morelos and Veracruz). These genetic distances were used to perform a Neighbor–Joining,17 Dendogram, using PHYLIP software v.3.6,18,19 comprising the programs GENDIST and NEIGHBOR. In addition, the proportion of Amerindian, European, African, and Asian specific genotypes were calculated based on the frequencies of ethnic specific HLA-A, HLA-B and HLA-DRB1 loci. The search of HLA origin was made whit the database available in www.allelefrequencies.net.
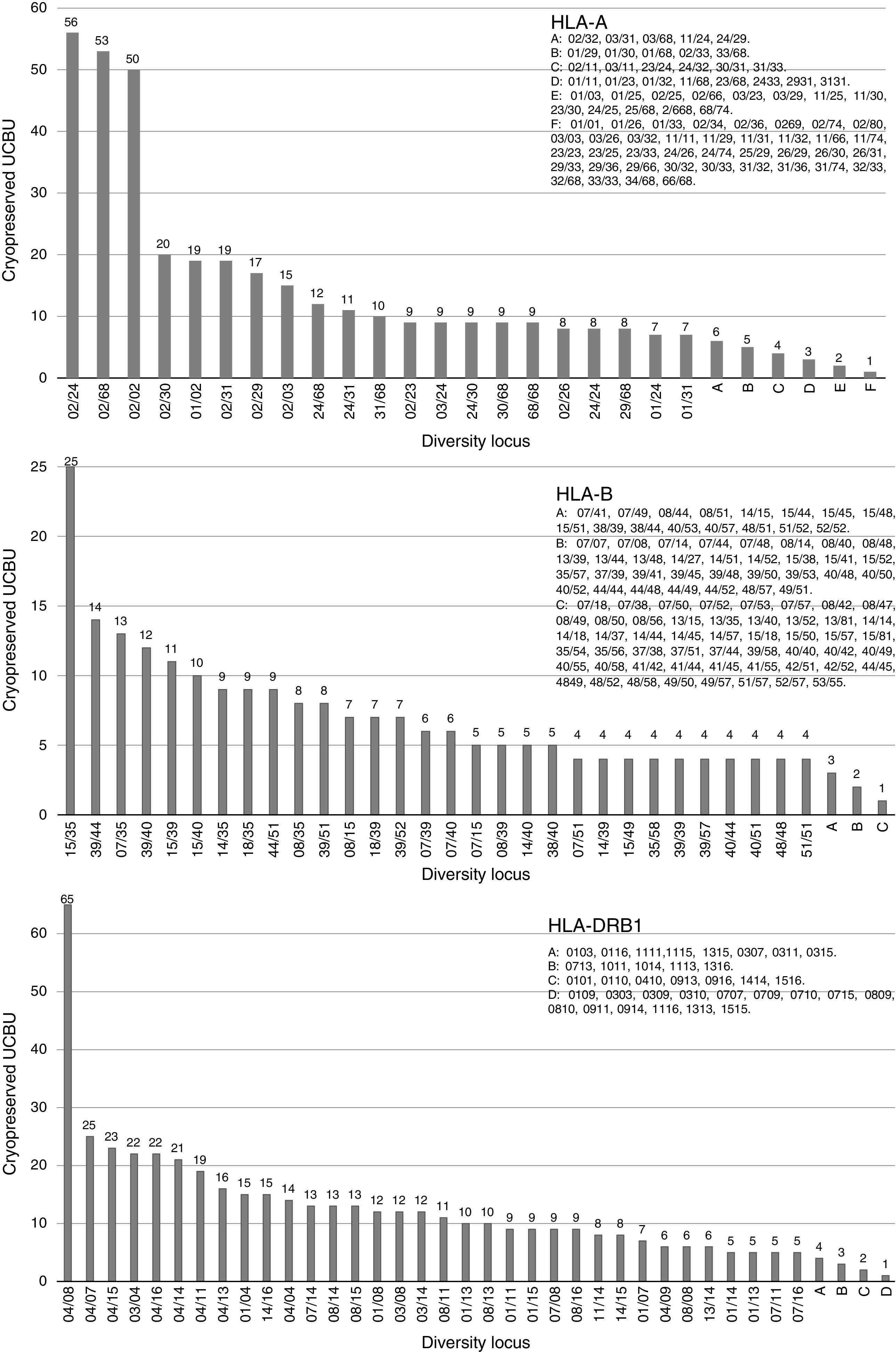
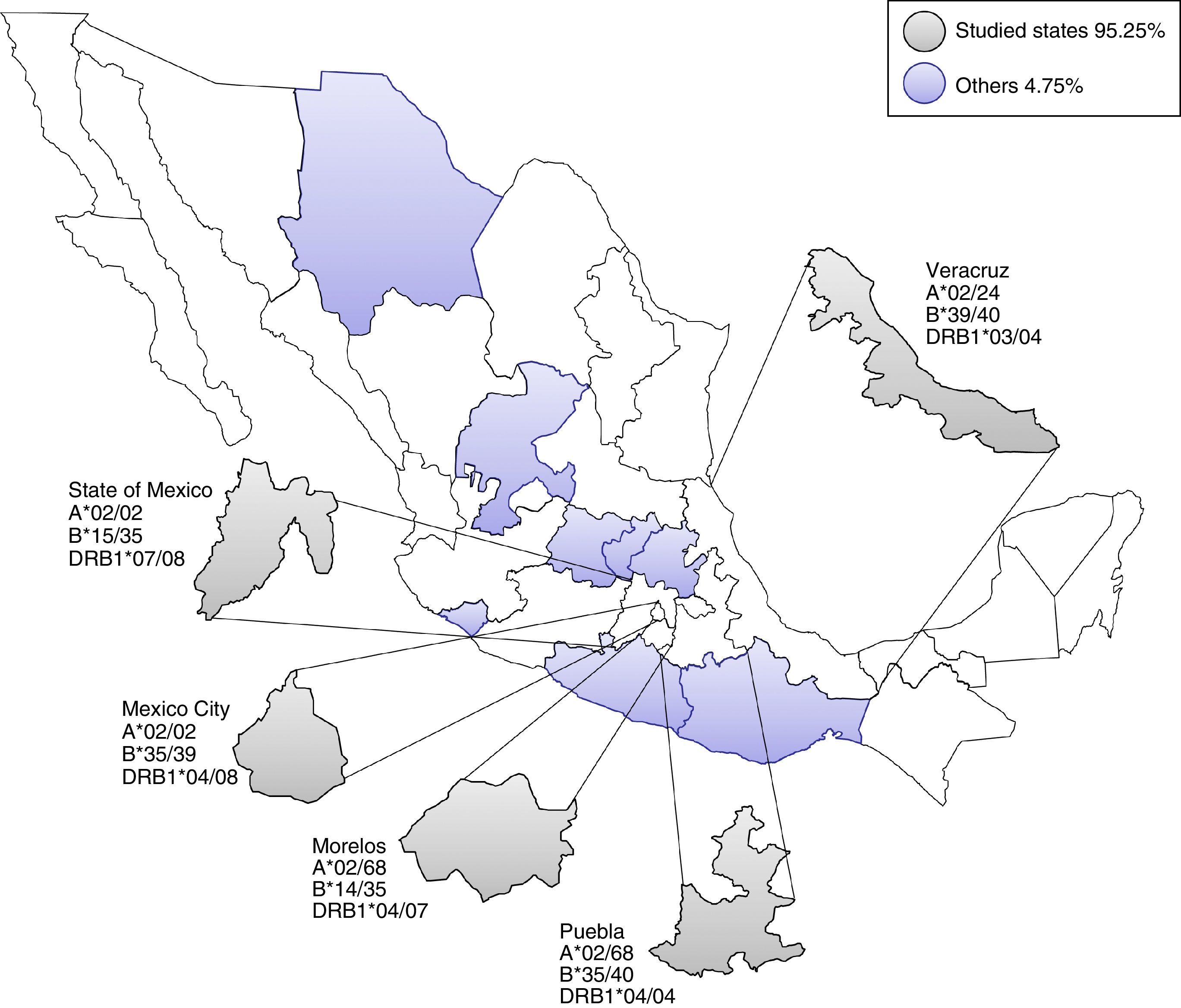
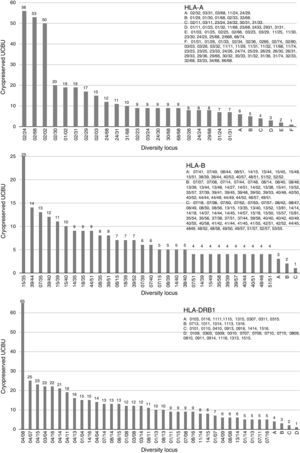
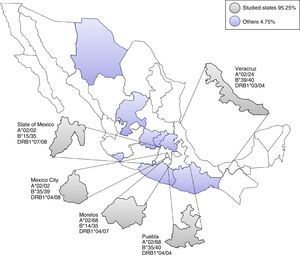
ResultsHLA distribution in Mexican UCB donorsHLA genotype frequencies data were estimated for 533 UCBU collected from 15 states of the Mexican Republic. A summary of the origin for the UCBU in the Mexican Republic is shown in Table 1. 46.5% (248) of the UCBU were obtained from Mexico City, 30.95% (165) State of Mexico, 8.06% (43) Puebla, 6.37% (34) Morelos and 3.37% (18) Veracruz. The remaining UCBU, 4.75% (25) were represented by other states in less proportion. Ninety-seven different HLA-A, one hundred sixty different HLA-B and sixty-nine different HLA-DRB1 loci were found in the UCBU analyzed. A complete list of the genotype frequencies data of HLA-A, HLA-B, and HLA-DRB1 loci are shown in Supplementary material. The most frequent locus (Fig. 1) were as follows: HLA-A: A*02/24 (10.5%), A*02/68 (9.9%), A*02/02 (9.3%), A*02/30 (3.7%), A*01/02 (3.5%), A*02/31 (3.5%); for the HLA-B locus, B*35/39 (4.6%), B*15/35 (4.6%), B*35/40 (3.3%), B*39/44 (2.6%), B*07/35 (2.4%), B*35/48 (2.2%), B*39/40 (2.2%) and for HLA-DRB1 locus, DRB1* 04/08 (12.2%), DRB1* 04/07 (4.7%), DRB1* 04/15 (4.1%), DRB1* 04/15 (4.3%), DRB1* 04/03 (4.3%) and DRB1* 04/14 (3.9%). The most frequent haplotype from the top five states and Mexico City (Mexico City, State of Mexico, Puebla, Morelos and Veracruz) analyzed are shown in Fig. 2.
Origin of cryopreserved Umbilical Cord Blood Units in the Cord Blood Bank from National Center of Blood Transfusion.
| State | UCBU collected n=533 |
|---|---|
| Mexico City | 248 |
| State of Mexico | 165 |
| Puebla | 43 |
| Morelos | 34 |
| Veracruz | 18 |
| Zacatecas | 9 |
| Hidalgo | 3 |
| Queretaro | 3 |
| Tlaxcala | 3 |
| Oaxaca | 2 |
| Chihuahua | 1 |
| Colima | 1 |
| Guanajuato | 1 |
| Guerrero | 1 |
| Yucatán | 1 |
Most frequent haplotype from the top five states UCBU collected in Mexican Republic. Ninety five point three percent of the UCBU was obtained from Mexico City, State of Mexico, Puebla, Morelos and Veracruz states (gray color). The remaining 4.7% of the UCBU was collected from other states (purple color).
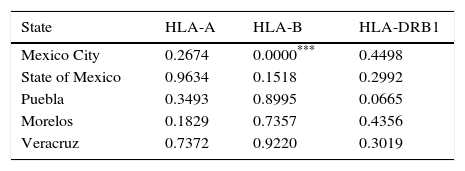
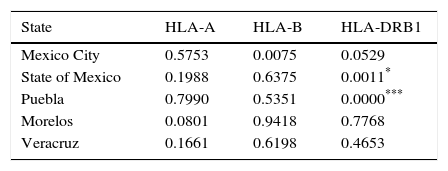
Hardy-Weinberg Test was performed using the top five populations and the three loci studied, populations are in equilibrium when locus HLA-A, HLA-B and HLA-DRB1 were evaluated separately, an exception is Mexico City population showing disequilibrium when locus HLA-B was evaluated (Table 2). So, a heterozygote excess proof was made with the same five populations and three loci studied (Table 3), showing that Mexico City present an excess of heterozygotes when HLA-B is evaluated, State of Mexico is higher significant for the same proof when is evaluated against HLA-DRB1 and Puebla shows the highest significance for excess of heterozygotes with the same locus.
P values in Hardy–Weingberg Test estimated by locus.
| State | HLA-A | HLA-B | HLA-DRB1 |
|---|---|---|---|
| Mexico City | 0.2674 | 0.0000*** | 0.4498 |
| State of Mexico | 0.9634 | 0.1518 | 0.2992 |
| Puebla | 0.3493 | 0.8995 | 0.0665 |
| Morelos | 0.1829 | 0.7357 | 0.4356 |
| Veracruz | 0.7372 | 0.9220 | 0.3019 |
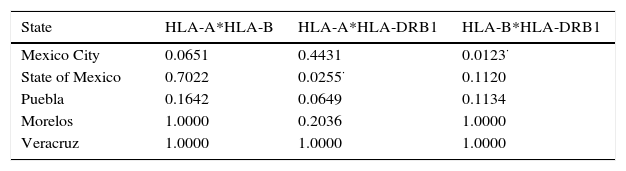
Globally, genotypic linkage disequilibrium showed that populations are independient between pairs of loci (Table 4), even though P values for linkage of Mexico City is slightly significant when HLA-B and HLA-DRB1 loci were evaluated paired; the same phenomenon occurs when HLA-A*HLA-DRB1 was evaluated for State of Mexico., meaning that those loci are segregating together in those populations, In order to follows the analyses, assumptions were made, in most populations (a) there's no-transmission of loci and (b) are in disequilibrium, allowing us to evaluate loci separately,
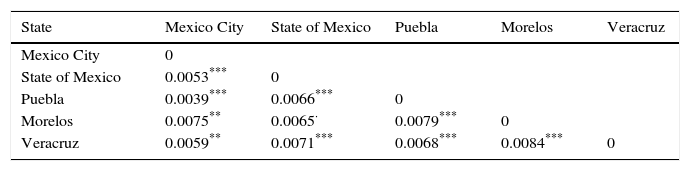
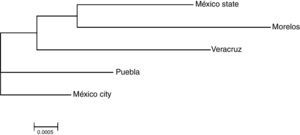
The genetic frequencies were used to calculate Cavalli-Sforza's genetic distances (Dc) between the top five Mexico regions were performed showing the genetic differences between them when compared through their genetic frequencies (Table 5). The shortest distance is that between Mexico City and Puebla (0.0039) and the largest is that between Veracruz and Morelos (0.0084), even though there's no clear relationships between states, in order to identify significant differences between UCBU of the Mexican Republic states (Mexico City, State of Mexico, Puebla, Morelos and Veracruz) an ANOVA test was performed, demonstrating that those populations of UCBU are significantly different from each other (P < 0.05). Which is more evident in the Neighbor-Joining Dendogram (Fig. 3) that shows the same effect with Puebla, forming a different branch with no root.
Genetic distances (Dc) among the top five states of the Mexican Republic with their significant differences between cryopreserved UCBU in the CBB from NCBT.
The present study describes the genotype frequencies of HLA Class I (A y B) and Class II (DRB1) in cryopreserved UCBU in the CBB at the NCBT. The genotype frequencies found show an important genetic diversity. Heterogeneity of the Mexican populations, where a stronger Caucasian component is preserved in the north of the country has been previously reported.20 The genotypic linkage disequilibrium showed that populations are independient between pairs of loci In Mexico City, a slightly significant linkage are showed when HLA-B and HLA-DRB1 loci were evaluated paired; the same phenomenon occurs when HLA-A*HLA-DRB1 was evaluated for State of Mexico, this perhaps due to the high proportion of mixing of genes as a result of high migration of the interior states of the Republic to these two cities. The Hardy-Weinbrg analysis revealed a significant deviation at locus HLA-B in Mexico City and at locus HLA-DRB1 in Estate of Mexico and Puebla, this was attributable to mostly heterozygous genotypes presents in this cities. The results demonstrate a combination of genotypes of varied ethnic origins; the majority of the allele groups found suggest Amerindian and European origins and in a lesser proportion Oriental and African. For HLA-A, all alleles that were found in the cryopreserved UCBU has been described in previous studies.21–23 The locus A*02/24 has the highest frequency; very common in Tarasco's and Han population in Southwest China,21,24 followed by locus A*02/68, very prevalent in Amerindian, Mestizo populations and Asian aborigines.25 The third most frequent locus in the studied UCBU is A*02/02, which confirms the Amerindian and Mestizo origins. Also the presence of alleles of Caucasian and African origins was detected in a lesser amount, in UCBU from Mexico City (A*66) and Puebla (A*33) and Veracruz (A*68) states. Thus, the genotype frequency profile generated for this gene shows a combination of high-frequency allele from Amerindian and Mestizo populations. Due to that the HLA-B has a greater polymorphism in the HLA system, greater genetic variability was identified in the UCBU studied (Fig. 1). For HLA-B, the B*35/39 and B*35/15 locus have the highest frequencies previously described in the Zapotec, Mixtec, Mixe and Tarasco population from the states of Oaxaca.21 The second most frequent locus in UCBU is B*35/40. The third frequent locus in UCBU is B*07/35. This combination of alleles represent more than 50% in all UCBU studied. These alleles have been previously described in Mexican populations, related to an Mestizo origin.26 It is important to note that HLA-B*35 is one of the predominant in Amerindians,4,27 and at the present study. The HLA-B*39 has been described before in the Mazatecans and other Mexican Amerindian populations.23 Results for HLA-B exhibit remarkable diversification of this agreeing with previous reported studies.21,26,27 At HLA-DRB1, the locus DRB1*04/08 is the most common in this locus, followed by DRB1*04/07 and DRB1*04/15. All of these HLA class II alleles have already been described in other Mexican populations and in population from northeast Mexico.22,23 HLA-DRB1*04 allele is relatively common in Mestizo population.28,29 The National Institute of Anthropology describes Mexican mestizos as individuals born in Mexico, descendant from the original inhabitants of the region and from individuals of Caucasoid, mainly Spaniards, and African origin, have a spanish-derived last name, and have a family of Mexican ancestors as far African as the third generation.25 The genetic distances analysis showed that the top five populations analyzed in this study (Mexico City, State of Mexico, Puebla, Morelos and Veracruz states) are significatively different from each other. These results are consistent with other studies that the Mexican Mestizo population is principally characterized by genotypes presents in Amerindian and Caucasian populations with a low frequency of Africans haplotypes. These observations may be influenced by the sample size, and interpopulation variations that may exist. The distribution of Caucasian as well as African admixture in Mestizos, however, is not homogeneous throughout the country. Several studies showed diverse levels of admixture depending on the region.22,23 Another factor that directly affects the genetic diversity is emigration and immigration. In summary, the genetic diversity of HLA gene identified concur with the tri-racial component existing in Mexican Mestizos reported previously. But there is a group of patients for which the NCBT cannot find a compatible UCBU because of the mixed ethnic origin. For example, the population of northern Mexico is mostly Caucasian. Most of the NCBT donors are of various ethnic origins, predominantly Amerindians and Caucasians; although some ethnic minorities like Oriental, African and pure Indian ethnics are not represented. Is important to note, that the sample size is considered a limitation in this study; since it is difficult to estimate the genotypic frequency at national level because most units of umbilical cord blood have been concentrated in the City and State of Mexico. On the other hand, the level of resolution of molecular techniques, does not allow a thorough analysis of the data. Allelic groups obtained by medium resolution techniques do not identify distance distributions, allele discrimination within a subgroup, even functional or structural variation. Based upon results obtained from this study, the NCBT is therefore establishing new agreements with the different states of the Mexican Republic to promote the donation of UCB in order to enrich the genetic diversity of UCBB in the CBB of this National Center.
Ethical disclosuresProtection of human and animal subjectsThe authors declare that no experiments were performed on humans or animals for this study.
Confidentiality of dataThe authors declare that no patient data appear in this article.
Right to privacy and informed consentThe authors declare that no patient data appear in this article.
Conflict of interestThe authors declare that they have no conflict of interests.