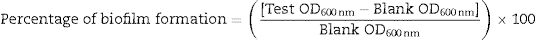Escalating burden of antibiotic resistance that has reached new heights present a grave concern to mankind. As the problem is no longer confined to clinics, we hereby report identification of a pandrug resistant Escherichia coli isolate from heavily polluted Delhi stretch of river Yamuna, India. E. coli MRC11 was found sensitive only to tobramycin against 21 antibiotics tested, with minimum inhibitory concentration values >256μg/mL for amoxicillin, carbenicillin, aztreonam, ceftazidime and cefotaxime. Addition of certain heavy metals at higher concentrations were ineffective in increasing susceptibility of E. coli MRC11 to antibiotics. Withstanding sub-optimal concentration of cefotaxime (10μg/mL) and mercuric chloride (2μg/mL), and also resistance to their combinatorial use, indicates better adaptability in heavily polluted environment through clustering and expression of resistance genes. Interestingly, E. coli MRC11 harbours two different variants of blaTEM (blaTEM-116 and blaTEM-1 with and without extended-spectrum activity, respectively), in addition to mer operon (merB, merP and merT) genes. Studies employing conjugation, confirmed localization of blaTEM-116, merP and merT genes on the conjugative plasmid. Understanding potentialities of such isolates will help in determining risk factors attributing pandrug resistance and strengthening strategic development of new and effective antimicrobial agents.
Pollution of the aquatic environment through anthropogenic activities, such as discharge of municipal sewage and untreated or partially treated industrial waste, creates means for selection, proliferation and dissemination of resistant traits among bacteria. Dissemination of resistance genes that diminishes the treatment options of infectious diseases has compromised human and animal health against multidrug resistant bacteria.1 Pollution of aquatic environments is common in developing countries like India, one such case occurs with the largest tributary of river Ganga – Yamuna, having its origin from Yamunotri Glacier of Uttar Kashi in Uttarakhand.2 It enters the capital territory Delhi at Palla village 15km upstream of Wazirabad barrage, that acts as a reservoir accounting for more than 70% of Delhi's water supply. Moreover, it is mainly the Delhi stretch of river Yamuna that receives high amount of discharge in terms of domestic sewage and industrial waste from urban centres and there are also various industrial settings established in and around the capital city.3 Providing a better niche for their establishment, pollution of aquatic environment selects bacteria by causing exchange of genetic determinants as well as acquisition of resistance traits crucial for it to thrive under the increasing pressure of pollutants. There are several reports pertaining to the dissemination of antimicrobial resistance genes, and it is known that their related genetic factors originate and are later transferred, from one bacteria to other bacteria present at a distant place.4–6
Development of resistance in bacteria makes it potent to compete with the sensitive ones, particularly in selective environments. Co-resistance to antibiotics and metals share common structural and functional strategies, conferred by chromosomal or mobile genetic elements.7,8 There is an increasing concern regarding potential of metal contaminated environment, acting as a pool, for sequestering antibiotic resistance genes, both in environmental and clinical isolates.9,10 Studies reporting assembly of resistance determinants on the same genetic element, insinuates towards the need to elucidate the role of polluted environment in acquisition of resistance in both pathogenic and non-pathogenic isolates of aquatic habitats.11–14 Exploring the resistance profiles among microbes inhabiting the aquatic ecosystem in urban areas, is of significant importance, as they directly or indirectly influence the sanitation outlook of the area. The presence of mercury and other heavy metals in water samples collected from river Yamuna has been reported in the studies of Sehgal et al.15 and Malik et al.16 Tolerance of mercury at different concentrations, suggests operation of different modes of detoxification encoded by genes located on mer operon among bacterial inhabitants of the polluted environment. Additionally, mer operon genes have been frequently observed to be genetically linked to antibiotic resistance genes.12,17 Taken together, investigation of mer operon genes as a representation of resistance to mercury, becomes pre-requisite to determine its role in selection and survival of bacterial isolates in the polluted environments. Thus, in the present study, cefotaxime resistant Escherichia coli isolates from Delhi stretch of river Yamuna were assessed for its susceptibility towards wide range of antibiotics and heavy metals. Genetic determinants imparting ESBL positive phenotype, conjugation frequency and biofilm formation in different media composition were studied for the isolate. Additionally, growth kinetics and susceptibility pattern to antibiotic and heavy metal either singly or in combination were also performed.
MethodologyIsolation and characterization of bacteriaUpon collection of the water samples from thirteen diverse locations – spread across Delhi stretch during March–April and September–October, 2012 and 2013 – bacterial screening was done by spreading 100μL of serially diluted samples on MacConkey agar plates supplemented with cefotaxime (4μg/mL). Non-duplicate bacterial colonies with distinct colony morphology were selected and subjected to characterization by IMViC test and 16S rRNA gene sequence analysis.
Antimicrobial susceptibility testThe antibiotic susceptibility test was done by disc diffusion method following Clinical and Laboratory Standards Institute18 guidelines. All isolates were subjected to drug susceptibility test against 21 antibiotics as described previously.19 Representing 13 different categories that includes cephalosporins, carbapenems, aminoglycosides and fluoroquinolones. Multiple Antibiotic Resistance (MAR) index was calculated, as described by Krumperman.20 Categorization of bacterial isolates into, multidrug resistant, extensively drug resistant and pandrug resistant, was strictly done on the basis of their resistance profiles following guidelines of Magiorakos et al.21 Minimum inhibitory concentration (MIC) of antibiotics was determined by broth micro-dilution method using Luria Bertani (LB) broth and E-test following CLSI guidelines with minor modifications in the culture medium. Similarly, resistance tests against heavy metals were determined by broth micro-dilution method by using different concentrations of heavy metal salts of cadmium, copper, chromium, mercury, lead and zinc. The cultures (96-well culture plates) were incubated at 37°C for 14–18h and Optical Density (OD) was measured at 600nm. The minimum concentration of each antibiotic and heavy metal salt inhibiting the growth of isolate was considered as its MIC. An isolate designated in this study as MRC11, showing broader resistance to different class of antibiotics and heavy metals, was selected for further studies.
Phenotypic screening for ESBL productionFollowing susceptibility test to 3rd generation cephalosporins [ceftazidime (CAZ), cefotaxime (CTX) and ceftriaxone (CTR)], selected bacterial isolates were screened for the production of extended spectrum β-lactamase enzyme(s) by Phenotypic Disc Confirmatory Test (PDCT) on Mueller-Hinton Agar plates as described earlier.19E. coli ATCC 25922 and Klebsiella pneumoniae ATCC 700603 were used as negative and positive controls, respectively.
Determination of ESBL and mer operon genesThe cefotaxime resistant isolate was screened for ESBL determinants (blaTEM, blaSHV and blaCTX-M) and carbapenem resistance genes (blaNDM-1, blaOXA-23 and blaOXA-48) along with the determinants of mer operon genes (merB, merP and merT), that confer resistance against mercury. Using gene specific primers (Table 1), PCR reactions were carried out as described earlier.19 Amplified PCR products were sequenced directly using automated DNA sequencer at Xcelris Labs Ltd., Gujarat, India. DNA sequence data was analyzed for homology using BLAST online search engine at NCBI (National Centre for Biotechnology Information).
Primer sequences used for gene amplification.
| Primer | Sequence | Product size (bp) | Reference |
|---|---|---|---|
| ID-F | 5′-GGCGGACGGGTGAGTAATG-3′ | 649 | [19] |
| ID-R | 5′-ATCCTGTTTGCTCCCCACG-3′ | ||
| TEM-F | 5′-ATGAGTATTCAACATTTCCGTGT-3′ | 861 | |
| TEM-R | 5′-TTACCAATGCTTAATCAGTGAGG-3′ | ||
| SHV-F | 5′ATGCGTTATATTCGCCTGTGTATTATCTCCC-3′ | 860 | |
| SHV-R | 5′-TTAGCGTTGCCAGTGCTCGATCAG-3′ | ||
| CTX-MF | 5′-SCVATGTGCAGYACCAGTAA-3′ | 480 | |
| CTX-MR | 5′-GCTGCCGGTYTTATCVCC-3′ | ||
| merPF | 5′-ATGAAGAAACTGTTTGCCTCC-3′ | 276 | |
| merPR | 5′-TCACTGCTTGACGCTGGACG-3′ | ||
| merTF | 5′-TTAATAGAAAAATGGAACGAC-3′ | 351 | |
| merTR | 5′-ATGTCTGAACCACAAAACGGG-3′ | ||
| merBF | 5′-ATGAAGCTCGCCCCATATATTTTAG-3′ | 667 | |
| merBR | 5′-TCACGGTGTCCTAGATGACATGG-3′ | ||
| NDM1F | 5′-GCATAAGTCGCAATCCCCG-3′ | 194 | This study |
| NDM1R | 5′-GGTTTGATCGTCAGGGATGG-3′ | ||
| OXA23F | 5′-GAAGCCATGAAGCTTTCTG-3′ | 200 | |
| OXA23R | 5′-GTATGTGCTAATTGGGAAACA-3′ | ||
| OXA48F | 5′-CCAATAGCTTGATCGCCCTC-3′ | 209 | |
| OXA48R | 5′-CCATAATCGAAAGCATGTAGC-3′ |
Localization of resistance genes was performed by conjugation studies followed by screening through PCR based amplification. ESBL producing test isolate was designated as donor, and a plasmid free, sodium azide resistant strain of E. coli (J53 AzR) sensitive to 21 antibiotics, served as recipient in the conjugation studies. Briefly, donor and recipient cells in a ratio of 1:2 were mixed in 5mL LB broth and incubated for 48h at 37°C without shaking. The trans-conjugants were selected on nutrient agar plates supplemented with cefotaxime (10μg/mL) and sodium azide (100μg/mL). The frequency of plasmid transfer was calculated with respect to the donor cells. An E. coli isolate positive for ESBL production was used as a control. In order to study co-transfer of multiple resistance markers in the conjugant, antibiotic profiling was performed against 21 antibiotics. Transfer of complete conjugative plasmid was confirmed from test isolate and trans-conjugant by plasmid isolation using alkaline lysis method, followed by size determination by performing agarose gel electrophoresis. Furthermore, the isolated plasmid from trans-conjugant was used as template for PCR to check the presence of ESBLs and mer operon genes.
Growth kineticsEffect of antibiotic and heavy metal, separately and also in combination, was studied by measuring growth parameters at regular intervals in nutrient media and subsequent plotting of values in form of growth curves. For analysis of the growth pattern, secondary culture of the isolate was setup in four culture flasks containing (a) LB broth, (b) LB broth supplemented with 10μg/mL cefotaxime, (c) LB broth supplemented with 2μg/mL HgCl2, and (d) LB broth supplemented with 10μg/mL cefotaxime+2μg/mL HgCl2. The cultures were grown aerobically till saturation, in an automated incubator shaker (Scigenics, India) set at 150rpm and 37°C. Similar experimental conditions were set up for the test isolate, its trans-conjugant, also for negative (E. coli strain ATCC 25922) and positive controls (E. coli isolate positive for ESBL production).
Biofilm formation assayBiofilm formation was investigated by crystal violet assay following protocol described by O’Toole and Kolter.22 Briefly, bacterial culture with OD600 of 0.1 was diluted in tryptic soy broth (TSB) by adding 100μL broth to 100μL culture (1:1 ratio). Polystyrene microtitre plates (96-well) were inoculated with 200μL of diluted culture per well and incubated at 37°C for 48h. Following this, cultures were discarded and wells were gently washed four times with 200μL of sterile phosphate buffer saline (PBS, pH 7.2) to remove loosely associated bacteria. Cells adhered to the wall were stained with 200μL of 0.1% (w/v) crystal violet at room temperature for 20min. It was followed by a wash with PBS. The crystal violet that stained the cells was solubilized in 200μL of 95% (v/v) ethanol. Samples were incubated for 20min at room temperature and biofilm formation was quantified by measuring OD600 in an ELISA reader (Thermo Scientific, MultiscanGo). Change in the amount of biofilm formation under different media composition was observed by supplementing TSB with cefotaxime (2μg/mL), mercuric chloride (0.2μg/mL) and their combination, and the wells containing TSB alone were used as blanks. The percentage of biofilm formation was calculated by the formula:
Taking a note of sensitivity to antibiotics, E. coli isolate known for ESBL production was used as a positive control and Pseudomonas aeruginosa ATCC 9027, exhibiting sensitivity to antibiotics along with biofilm formation capability, was used as a negative control, in assessing the behaviour of the test isolate.
Binary toxicity assayThe multidrug resistant isolate was further subjected to combined toxic effect of heavy metals and antibiotics. The petriplates containing MHA supplemented with different concentrations of heavy metals was inoculated and spread with the bacterial culture. Combined toxicity was determined by measuring the zone of inhibition after placing antibiotic discs at appropriate distance and incubation for 12–14h at 37°C. On the basis of susceptibility, four antibiotics with three different modes of action: amikacin (aminoglycoside), ciprofloxacin (fluoroquinolone), cefoxitin (chephamycin) and imipenem (carbapenem), were included in this study. Each antibiotic was used at four different concentrations, in combination with six heavy metal salts [CdCl2 (175, 75, 50, 5mg/L), CuCl2 (250, 100, 50, 5mg/L), CrO3 (125, 30, 15, 5mg/L), HgCl2 (15, 7.5, 1, 0.1mg/L), Pb(CH3COO)2 (250, 125, 75, 10mg/L) and ZnSO4 (500, 250, 100, 50mg/L)]. The concentrations of heavy metals were selected on the basis of MIC obtained for each metal.
ResultsThe bacterial isolateA total of 75 non-duplicate E. coli isolates were collected from water samples in heavily polluted Delhi stretch of river Yamuna. Screening for ESBL production revealed 34 E. coli isolates to be positive for ESBL production. Out of these ESBL positive E. coli isolates, an isolate (annotated as MRC11) collected on March 26, 2013 from Old Railway Bridge, 100–150m downstream of Civil Mill Drain falling into the river (77°14′51.42″ E and 28°39′41.77″ N), displayed high incidences of resistance, therefore it was chosen for MDR studies against wide range of antibiotics and heavy metals. In addition to the identification by assessment of the biochemical parameters, recognition based on 16S ribosomal RNA gene sequence, confirmed the isolate to be an E. coli (NCBI Accession # KT428597).
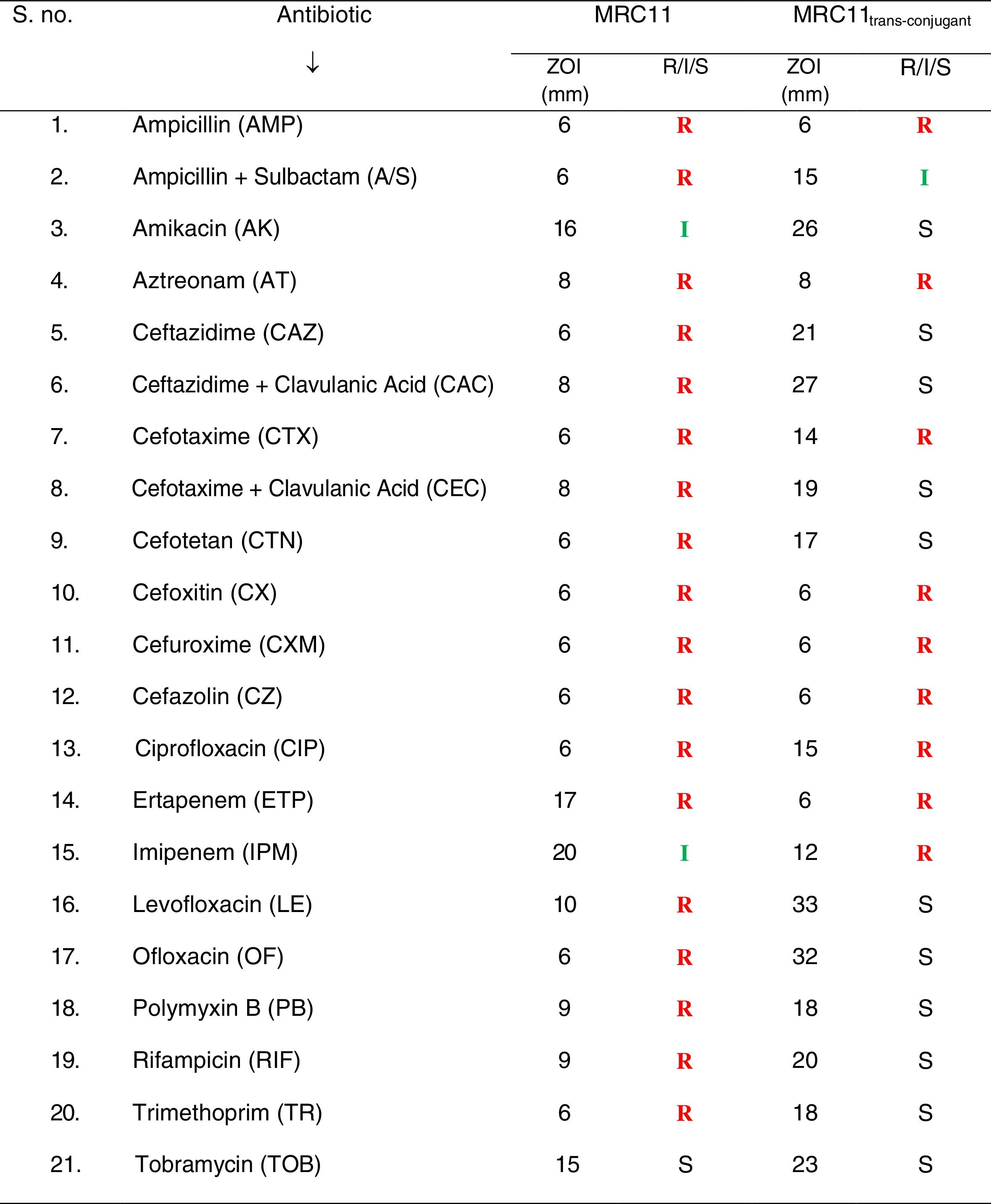
Antimicrobial susceptibilityBy checking susceptibility of the bacterial isolates against antibiotics, it was observed that E. coli MRC11 exhibits resistance against broad range of antibiotics representing β-lactams, monobactams, fluoroquinolones, polymyxins, rifampicins and trimethoprim (Table 2). Phenotypic determination of ESBL production revealed complete absence of zone of inhibition for third generation cephalosporins, individually, as well as in combination with the inhibitor i.e. clavulanic acid. The isolate was found to be sensitive to tobramycin and amikacin. In summary, resistance displays peaks for amoxicillin and carbenicilin (MIC >512μg/mL) followed by aztreonam, cefotaxime and ceftazidime (MIC >256μg/mL), ofloxacin (32μg/mL) and ciprofloxacin (30μg/mL), respectively (Table 3). The isolate showed complete resistance to ofloxacin and trimethoprim, comprising choice of drugs available for the treatment of UTI infections. Besides, MIC value of 2μg/mL (susceptible range) for amikacin, ZOI was observed to be in intermediate category (16mm). Higher MAR index (0.95) for the E. coli MRC11 gives an indication of its portrayal as a therapeutic challenge. Simultaneously, by looking at the resistance against different heavy metals, it was observed that E. coli MRC11 exhibits a very high resistance profile for metals, even higher than the prescribed permissible limit which is considered tolerable for humans (Table 4).
Analysis of DNA sequence data revealed presence of two different variants of blaTEM (blaTEM-1 and blaTEM-116; NCBI Accession # KU376497 and KT428598) gene in E. coli MRC11 that is credited towards contribution of resistance to β-lactams. As amplification could not be achieved for other two ESBL (blaSHV and blaCTX-M) genes and carbapenem resistance imparting genes (blaNDM-1, blaOXA-23 and blaOXA-48), it was concluded that the isolate is devoid of them. Presence of mer operon (merB, merP and merT) genes associated in imparting resistance against organic as well as inorganic mercury, gives an indication about the presence of both broad and narrow spectrum mer operon in the isolate.
Plasmid profilingConjugation efficiency of the magnitude of 3.5×10−2 and 6.1×10−5 trans-conjugant per donor cell was observed for E. coli MRC11 and control, respectively. Such high conjugation efficiency gives an indication of higher possibility of transfer of plasmid borne genes. Successful transfer of resistance determinants leads to the transformation of sensitive recipient into well acquaint resistant phenotype (Table 2). An ∼22kb plasmid DNA isolated from trans-conjugant confirms complete transfer of conjugative plasmid via conjugation. PCR showed positive amplification corresponding to blaTEM-116 along with mer operon (merP and merT) genes from trans-conjugant plasmid. The trans-conjugant was resistant against cephalosporins, monobactams, carbapenems and ciprofloxacin. Higher MIC values of cephalosporins (>256μg/mL), carbenicillins (>512μg/mL) and ciprofloxacin (5μg/mL) demonstrate high resistance range of the trans-conjugant (Table 3).
Growth studiesGrowth kinetic studies reflect the effect of antimicrobials (antibiotics and heavy metals), either alone or in combination, on the growth pattern of E. coli MRC11. Growth of E. coli MRC11 under normal conditions showed a lag phase of about 2h followed by a sharp increase in biomass during the log phase. On supplementing media with cefotaxime, no significant change in growth was observed for the isolate (Fig. 1A and B). However, supplementing media with mercuric chloride, an extended lag phase of 9h was observed for both test isolate and its trans-conjugant. Contrarily, to the expected extension in lag phase following supplementation of both cefotaxime and mercuric chloride, a decline in the lag phase by about 2h (a net lag phase of 7h) was observed, which may be attributed to the resistance determinants conferring co-resistance (Fig. 1A). However, growth in the case of positive control (E. coli ESBL+) was compromised following treatment with mixture of cefotaxime and mercuric chloride (Fig. 1C). Compared to this, negative control displays no growth patterns after the application of cefotaxime and mercuric chloride, either alone or in combination (Fig. 1D).
Growth pattern of bacteria under different culture conditions. (A) Isolate E. coli MRC11, (B) isolate E. coli J53 trans-conjugant, (C) positive control (i.e., isolate resistant to β-lactams and heavy metals) and (D) sensitive control E. coli ATCC 25922. The X axis depicts time in hours where as Y axis represents bacterial growth presented in terms of OD at 600nm.
Biofilm formation in nutrient media was around 59% in E. coli MRC11 and 79% in P. aeruginosa ATCC 9027 (Fig. 2). When assessed under different medium conditions, compared to the control, biofilm formation was found to follow a different pattern for E. coli MRC11 isolate. Maximum biofilm formation in the medium supplemented with mercuric chloride was seen in E. coli MRC11, also biofilm formation for the control isolate is maximum in the media which is supplemented with mercuric chloride and cefotaxime. The growth patterns of the biofilm formation for control is, higher in media supplemented with mixture of mercuric chloride and cefotaxime, which is followed by the growth seen in presence of mercuric chloride (68%) and the least growth (65.6%) occurs in the presence of cefotaxime alone. However, highest growth of the biofilm can be seen in the media supplemented with metal alone, which followed by the growth in media supplemented with cefotaxime (56.2%) and the least growth is seen in the presence of mercuric chloride and cefotaxime (Fig. 2).
Dual resistance towards antibiotics and heavy metalsKeeping in mind that the treatment with metal and antibiotics effects bacterial susceptibility, combined toxicity assays for E. coli MRC11 were performed. The detailed results of the combined toxicity test are summarized in Fig. 3. E. coli MRC11 isolate which showed intermediate phenotype towards amikacin, exhibits sensitive phenotype in presence of cadmium, copper and chromium. Increase in the zone of inhibition was observed when amikacin and copper, both were used together, at a concentration of 5mg/L and 50mg/L, respectively. While as no such change in the zone diameter was observed at 100mg/L and 250mg/L concentration, which gives a clear indication about their synergistic effect towards E. coli MRC11. In a similar manner, combination of amikacin with zinc showed increase in its activity as observed by the increase of zone diameter at 500mg/L concentration, with little or no change at the lower values. Contrary to this, mercury at a concentration of 1mg/mL reduced the zone diameter from 16mm to 14mm, thereby indicating that the two hold antagonistic effect.
Effect of heavy metal on antibiotic resistance pattern of E. coli MRC11. R (resistant), I (intermediate) and S (sensitive) category as per CLSI criteria were labelled on each column. WM represented sensitivity in absence of heavy metal; C1, C2, C3 and C4 were concentrations of heavy metals in the medium, that were, Cd (175, 75, 50, 5mg/L), Cu (250, 100, 50, 5mg/L), Cr (125, 30, 15, 5mg/L), Hg (15, 7.5, 1, 0.1mg/L), Pb (250, 125, 75, 10mg/L) and Zn (500, 250, 100, 50mg/L) respectively. NG showed no growth of E. coli MRC11 isolate at that particular concentration of heavy metal.
Similarly, combination of lead (75mg/L) with imipenem showed increase in zone diameter; that shifted the activity of E. coli MRC11 towards sensitive range. Zinc in combination with imipenem, showed a positive impact at 500mg/L, with little or no effect at 250mg/L, 100mg/L and 50mg/L. On one side, where imipenem showed positive correlation with lead and zinc, no differences were observed at varying concentrations of chromium and mercury. Higher concentrations of cadmium and copper have an antagonistic effect when used in combination with imipenem. The susceptibility pattern changed from sensitive to resistant in case of cadmium and from sensitive to intermediate for copper. Additionally, E. coli MRC11 isolate displayed complete phenotypic resistance to different concentrations of cadmium, copper, chromium, mercury, lead and zinc, when used in combination with ciprofloxacin and cefoxitin.
DiscussionSheer imprudence in the use of antibiotics has resulted in the emergence and dissemination of resistance determinants. Recognized as a major challenge in the healthcare systems, bacteria have developed mechanisms that render antibiotics useless against infections. Polluted environment exacerbates the situation by creating a selective pressure favouring acquisition of resistance genes and point mutations that leads to the development of newer gene variants with broader activity, ultimately increasing the resistance burden in different environmental conditions.19,23,24 One such location identified as Delhi stretch of river Yamuna, representing only 2% of the total catchment area of the river, gets discharge from around 22 sewage and 28 industrial clusters.2 This stretch is heavily eutrophicated at several locations, where concentration of almost all major heavy metals and antibiotics is well above the maximum permissible limits.16,25 Rather than acting as a graveyard for bacterial isolates where multiple antimicrobial agents work simultaneously, the polluted river Yamuna is crammed with isolates harbouring clusters of resistance genes.
Extended spectrum β-lactamase enzymes are reported as the main contributing factors to high level of resistance observed among majority of the multidrug resistant bacterial isolates. As such, rapid emergence of multidrug resistance among opportunistic pathogens that cause endocarditis, bacteraemia, sepsis, urinary tract and other infections, has now grown to an alarming condition, troubling clinicians treating them.26 Associated with urinary tract infections (UTIs), E. coli, which predominates among members of Enterobacteriaceae, seems to be a true community ESBL-producing pathogen.27,28 Delhi stretch of river Yamuna is crammed with multidrug resistant E. coli isolates, out of which 45% were ESBL producers. Among these, one isolate, E. coli MRC11 exhibits resistance against 20 antibiotics tested. Having an MAR index of 0.95 and high MIC values (>256μg/mL) to different antibiotics, consortium of available drugs seems inadequate in dealing with its infections. Similar to study carried out in Thailand where Pornsinchai et al. reported co-presence of blaTEM-1 and blaTEM-116 genes in clinical isolates of E. coli and K. pneumoniae, E. coli MRC11 isolate was found to harbour two different variants of blaTEM gene.29 Carbapenems being a choice of drug against ESBL producing isolates,30E. coli MRC11 exhibits resistance against ertapenem and imipenem. Absence of amplicon for blaNDM-1, blaOXA-23 and blaOXA-48 genes suggest presence of other determinants imparting carbapenem resistance. Similar to study of Rath et al. reporting prevalence of amikacin resistance among nosocomial and community acquired E. coli isolates, E. coli MRC11 in our study was seen to be resistant to amikacin.31 Fluoroquinolones constitute the most widely used anti-bacterials that are effective in treating UTIs caused by E. coli.32 Wide spread resistance to levofloxacin and ofloxacin in E. coli MRC11 often renders fluoroquinolones of no use. Resistance against one or more agents from thirteen different classes of antibiotics portrays pandrug resistance phenotype to E. coli MRC11 isolate. This is in concordance with the studies that advocate high incidence of multidrug resistance among bacterial isolates collected from water streams influenced by industrial pollution and municipal sewage.33,34
Clustering of the resistance genes and their mobilization through horizontal gene transfer armours bacteria with resistance against wide range of antibiotics.35,36 Complying with the reports, trans-conjugant of E. coli MRC11 that apprehends presence of resistance determinants on conjugative plasmid exhibits resistance to wide variety of antibiotics, in addition of having ESBL phenotype possibly attributed to presence of other resistance genes that are not examined in this study. Moreover, in our work it was observed that, trans-conjugant of E. coli MRC11 carries mer operon (merP and merT) genes. These results are in corroboration with the work of McIntosh et al. reporting genetic linkage between mer operon with that of antibiotic resistance genes.17 In combined toxicity test, amikacin in amalgamation with cadmium chloride and chromium (III) oxide showed synergistic effect possibly due to chemical reaction between the effective concentrations of heavy metals and antibiotics, and/or presence of heavy metal that elicit stress response, impairing protein (enzyme) synthesis, that impart resistance to the bacteria. Contrarily, when antimicrobial activity of the combination (antibiotics plus metal) is no more equal to or greater than parent substance, and/or coagulation occurs, it is considered as antagonistic effect as observed in the case of imipenem with cadmium chloride (175mg/L) and zinc (50mg/L), and amikacin with mercuric chloride (1mg/L) combinations. These results are complying with the observations of studies carried out by Zhou et al.37 that reports changed susceptibility of Psudomonas fluorescens to antibiotics when co-exposed to heavy metals.
Knowing the fact that blaTEM genes impart resistance to β-lactams, growth of the isolate E. coli MRC11 under study was found insensitive to cefotaxime (10μg/mL). Accordingly, E. coli MRC11 transformant harbouring resistance genes on plasmid follows almost similar growth pattern. While, presence of heavy metal in media reduces the microbial activity to a large extent, thereby increases lag phase, later replenishing the growth possibly due to co-resistance or detoxification strategies generated in the cell by the proteins encoded by the set of mer operon genes. Similar growth patterns, without any effect of antibiotic in medium, on the growth kinetics of antibiotic resistant bacteria, were also demonstrated by Drummond et al.38 Also, no significant increase in biofilm formation was observed in presence of antimicrobial agent for E. coli MRC11, compared to control isolate. The plausible explanation for these observations could come from the fact that potential expression of the ESBL gene on conjugative plasmid, might confer resistance to cephalosporins-resulting in well adapted phenotype – so that it can function or grow in a normal manner, without any effect on the growth pattern and also without significant induction of biofilm formation. Taken together, detection of E. coli MRC11 in aquatic environment presents a risk factor for dissemination of resistance genes to co-inhabitants including potential pathogens which pose serious threat to mankind. Understanding the potentialities of multiresistant isolates to withstand effect of antimicrobials will help in strategic development of new and more effective antimicrobial agents so as to “reset the clock” for resistance level and infection control. To achieve this, further studies are required to evaluate the role of polluted environment in resistance spread and to determine the factors responsible that attribute bacteria with the pandrug resistant phenotype.
Conflicts of interestAuthors declare that no conflict of interest exist with this publication.
Mudsser Azam acknowledges Council of Scientific & Industrial Research (CSIR), India for financial support as Senior Research Fellowship (09/466(0136)/2011-EMR-I).