Multiple genetic and environmental factors interact to determine an individual’s predisposition to non-alcoholic fatty liver disease and its phenotypic characteristics. Association studies have found a number of alleles associated with the development of non-alcoholic steatohepatitis. Our aim was to investigate whether multiple risk-associated alleles may be present in affected monozygotic twins, indicating underlying genetic predisposition to non-alcoholic steatohepatitis. We determined the genotype of 14 candidate gene polymorphisms (at 11 unlinked loci) in a set of monozygotic twins who presented with cirrhosis within 18 months of each other. Genotyping revealed multiple single nucleotide polymorphisms at 9 independent loci in genes PNPLA3, APOC3, GCKR, TRIB1, LYPLAL1, PPP1R3B, COL13A1, and EFCAB4B, previously implicated in contributing to non-alcoholic steatohepatitis pathogenesis. In conclusion, this case series illustrates the potential cumulative effect of multiple polymorphisms in the development and potential progression of a complex trait such as NASH cirrhosis.
Non-alcoholic fatty liver disease (NAFLD) has been considered highly heritable.1 However, as with any complex trait, multiple genetic and environmental factors interact to determine an individual’s predisposition to NAFLD and its phenotypic characteristics such as histological severity and disease progression. We describe identical twins presenting with cirrhosis secondary to non-alcoholic steatohepatitis (NASH) within 18 months of each other. A number of single nucleotide polymorphisms (SNPs) have been identified in candidate gene and genome-wide association studies (GWAS) as being associated with liver fat content, NAFLD/NASH.2-7 We deter-mined the genotype of the twins at these loci with an aim to elucidate an underlying genetic predisposition to their liver disease.
Material And MethodsThe study protocol conforms to the ethical guidelines of the 1975 Declaration of Helsinki as reflected in a priori approval by the East Midlands Research Ethics committee and both patients provided written informed consent for genetic testing and publication. Both cases had a negative liver screen (hepatitis A, B, C, and E serology, autoantibodies, serum copper, caeruloplasmin, ferritin and alpha1 antitrypsin levels), neither had abused alcohol, though both had insulin resistance (IR) (Table 1).
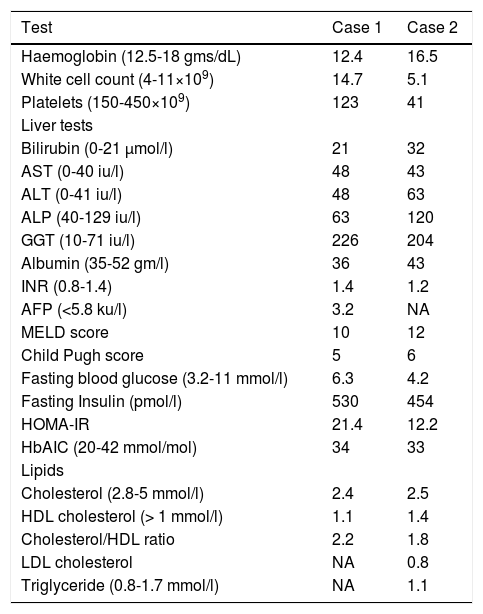
Laboratory data at presentation in the two cases.
| Test | Case 1 | Case 2 |
|---|---|---|
| Haemoglobin (12.5-18 gms/dL) | 12.4 | 16.5 |
| White cell count (4-11×109) | 14.7 | 5.1 |
| Platelets (150-450×109) | 123 | 41 |
| Liver tests | ||
| Bilirubin (0-21 μmol/l) | 21 | 32 |
| AST (0-40 iu/l) | 48 | 43 |
| ALT (0-41 iu/l) | 48 | 63 |
| ALP (40-129 iu/l) | 63 | 120 |
| GGT (10-71 iu/l) | 226 | 204 |
| Albumin (35-52 gm/l) | 36 | 43 |
| INR (0.8-1.4) | 1.4 | 1.2 |
| AFP (<5.8 ku/l) | 3.2 | NA |
| MELD score | 10 | 12 |
| Child Pugh score | 5 | 6 |
| Fasting blood glucose (3.2-11 mmol/l) | 6.3 | 4.2 |
| Fasting Insulin (pmol/l) | 530 | 454 |
| HOMA-IR | 21.4 | 12.2 |
| HbAIC (20-42 mmol/mol) | 34 | 33 |
| Lipids | ||
| Cholesterol (2.8-5 mmol/l) | 2.4 | 2.5 |
| HDL cholesterol (> 1 mmol/l) | 1.1 | 1.4 |
| Cholesterol/HDL ratio | 2.2 | 1.8 |
| LDL cholesterol | NA | 0.8 |
| Triglyceride (0.8-1.7 mmol/l) | NA | 1.1 |
Genomic DNA was prepared from whole blood using Flexigene DNA kit (Qiagen, Germany). Genotypes were determined by sequencing (Source Bioscience, UK) PCR products generated using the following primer pairs (Sigma-Aldrich, UK):
- •
rs738409: PNPLA3_1F, GAGCCAACAACCCTTGGTCCTGTCTG and PNPLA3_1R, GCTGCCCGGGTAGCCTGGAAATAG (546bp).
- •
rs6006460: PNPLA3_F, GTGTCTCTGGCTGTGACGGCACC and PNPLA3_3R, TCACTACACAGCAATGCGGAGGTA (454bp).
- •
rs2854116 and rs2854117: APOC3_1F, GAAGGTGAACGAGAGAATCAGTCCTG and APOC3_1R, GCCTCGGGCCCATCTCAGCCTTTCACACTG (513bp).
- •
rs2385114: TRIB_1F, ACAGACCTTTGAGAGGAAAAGGAATTGACC and TRIB_1R, GCTGGTGCCCACACTGATACATGC (548bp).
- •
rs7800894: GCKR_1F, GAGACATAGTCTCACTCTGTTGCCCAGG and GCKR_1R, CCCAGCGTTTAAATGTTAGGGCAATAGATGTAAACC (588bp).
- •
rs1260326: GCKR_2F, GGGTCTTAGGGTACCTGCTCAGAGG and GCKR_2R, GGTAACCCATGACCTTGCCCAGC (546bp).
- •
rs2228603: NCAN_1F, GCCTCTGAATCAGGCCCTCTCTCC and NCAN_1R, GGTGACATTCAGTGGGTGGGCTCC (520bp).
- •
rs2645424: FDFT_1F, CCTCTCAAAAGAAGAGGAAGGGCTTGG and FDFT_1R, GAGACACCACACTTAGCCTCACC (403bp).
- •
rs1227756: COL13A_1F, ACCATCCATGGCTCCTCAGTGC and COL13A_1R, GGCGGAGATGGCAAGGAAGCTG (592bp).
- •
rs887304: EFCAB4B_1F, CCCATCAGTAAATTCCTGTGAATGAACTGCCCTACTCACC and EFCAB4B_1R, CTTGGTCCCCAGTGCAGTGTTTGGACCA (274bp).
- •
rs4240624: PPP1R3B_2F, CCAGGCTGGTCTCGATCCCCTTGG and PPP1R3B_2R, ACCCAAACGGAGTGCCAGTACCACG (525bp).
- •
rs58542926: TM6SF2_F1 CAGGCATGAGCCACCGCACC and TM6SF2_R2 ACAGATGTCCAGCAGGGTTC (330bp).
- •
rs12137855: LYPLAL_1F, TAAAAGTTTACATGCCCTCCTCTTATAGG and LYPLAL_1R, GCCCATGTAGTTGTTACAGACACTGG (430bp).
Phusion polymerase and HF buffer (New England Biolabs, UK) was used to amplify each region from the genomic DNA templates and products were gel purified (Machery Nagel, Germany).
ResultsCase 1A 69 year old Caucasian male was referred to the haematologists in July 2010 with thrombocytopenia (platelet count of 53 × 109/L). Bone marrow examination was suggestive of consumptive thrombocytopenia with normal cytogenetics. A subsequent liver ultrasound indicated cirrhosis and he was referred to Hepatology services in November 2010. He drank about 14 units of alcohol/week, and had mild hypertension, there being no significant family or drug history. On examination besides palmar erythema there were no other stigmata of chronic liver disease. His BMI was 27.2 with a waist hip ratio of 0.97, though there had been a 14 kg unintentional weight loss over the last six months. Initial laboratory tests are shown in table 1. A liver screen was negative and liver ultrasound suggested cirrhosis with a splenomegaly of 14.7 cm. A percutaneous liver biopsy performed on 23rd March 2011 confirmed cirrhosis with patchy steatosis involving 15% of the hepatocytes. He did have a variceal bleed in March 2011 requiring banding and developed grade 2 hepatic encephalopathy that has responded to medical therapy. More recently he has been diagnosed with hepatocellular cancer (HCC).
Case 2A 71 year old Caucasian man presented to our institute in November 2012 with hematemesis and melena. He had drunk alcohol (<16 units/week) for about 10 years aged 40-50 years. Of note his identical twin brother had been diagnosed with cirrhosis in 2010 (as above).There was no significant past medical history and he was on no medication. On examination he had liver palms, multiple spider angiomata, a firm 3 cm hepatomegaly and moderate ascites. His BMI was 28.4, though he had had an unintentional 7 kg weight loss in the last four months. Initial laboratory tests are shown in table 1. An urgent gastroscopy revealed grade two oesophageal varices (with stigmata of recent bleeding) that were banded. A liver screen (see methods) was negative and transient elastography (TE) (Echosens 502 Touch) confirmed cirrhosis (liver stiffness measurement (LSM) 15.9 kPa) with controlled attenuation parameter (CAP) of 266 dB/m. A triple phase CT scan of the liver revealed a 6 cm HCC in segment 3. He was referred to the regional tertiary centre where he underwent successful transarterialchemoembolization (TACE).
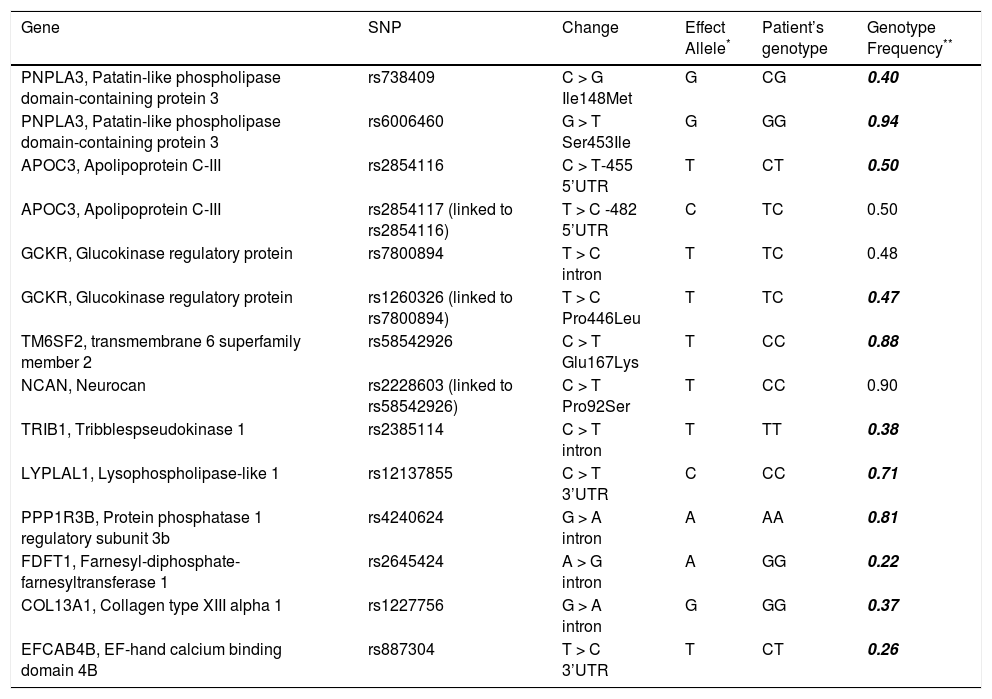
Patient genotypesThe genotyping results are listed in table 2 along with the effect alleles and the expected frequencies for these genotypes. This analysis revealed that both patients were heterozygotes for: PNPLA3 rs738409 (Ile148Met); APOC3 rs2854116; APOC3 rs2854117 (which is closely linked to rs2854116); GCKR rs7800894, and EFCAB4B rs887304 indicating that they had a single ‘risk’ allele at 4 independent genetic loci. We also found heterozygosity for a functional GCKR variant (Pro446Leu) rs1260326, which has been shown to beclosely linked to rs7800894.8 The twins were homozygotes for the ‘risk’ allele at: PNPLA3 rs6006460 (encoding Ser453); TRIB1 rs2385114; LYPLAL1 rs12137855; PPP1R3B rs4240624 and COL13A1 rs1227756.The ‘risk’ allele was absent for FDFT1 rs2645424, NCAN rs2228603 (encoding Pro92) and the linked TM6SF2 rs58542926 (encoding Glu167). The probability of having this allele combination at the 11 separate loci (when only one of closely linked polymorphisms is included) implicated in disease development, by chance would be 0.00036 assuming Hardy-Weinberg equilibria based on the population minor allele frequencies listed in the Ensembl database.
SNP details and genotype of twins.
| Gene | SNP | Change | Effect Allele* | Patient’s genotype | Genotype Frequency** |
|---|---|---|---|---|---|
| PNPLA3, Patatin-like phospholipase domain-containing protein 3 | rs738409 | C > G Ile148Met | G | CG | 0.40 |
| PNPLA3, Patatin-like phospholipase domain-containing protein 3 | rs6006460 | G > T Ser453Ile | G | GG | 0.94 |
| APOC3, Apolipoprotein C-III | rs2854116 | C > T-455 5’UTR | T | CT | 0.50 |
| APOC3, Apolipoprotein C-III | rs2854117 (linked to rs2854116) | T > C -482 5’UTR | C | TC | 0.50 |
| GCKR, Glucokinase regulatory protein | rs7800894 | T > C intron | T | TC | 0.48 |
| GCKR, Glucokinase regulatory protein | rs1260326 (linked to rs7800894) | T > C Pro446Leu | T | TC | 0.47 |
| TM6SF2, transmembrane 6 superfamily member 2 | rs58542926 | C > T Glu167Lys | T | CC | 0.88 |
| NCAN, Neurocan | rs2228603 (linked to rs58542926) | C > T Pro92Ser | T | CC | 0.90 |
| TRIB1, Tribblespseudokinase 1 | rs2385114 | C > T intron | T | TT | 0.38 |
| LYPLAL1, Lysophospholipase-like 1 | rs12137855 | C > T 3’UTR | C | CC | 0.71 |
| PPP1R3B, Protein phosphatase 1 regulatory subunit 3b | rs4240624 | G > A intron | A | AA | 0.81 |
| FDFT1, Farnesyl-diphosphate-farnesyltransferase 1 | rs2645424 | A > G intron | A | GG | 0.22 |
| COL13A1, Collagen type XIII alpha 1 | rs1227756 | G > A intron | G | GG | 0.37 |
| EFCAB4B, EF-hand calcium binding domain 4B | rs887304 | T > C 3’UTR | T | CT | 0.26 |
The sequences obtained for both patients were identical for all the polymorphisms tested and for SNPs in the flanking regions sequenced (totalling 5232 bp and containing an additional 64 SNP sites).
DiscussionWe describe two cases (monozygotic twins) who presented with cirrhosis within 18 months of each other. In the first case, percutaneous liver biopsy revealed cirrhosis with background hepatic steatosis. In the second case TE suggested cirrhosis (15.9 kPa) with a CAP of 266 dB/m (indicative of hepatic steatosis).9 Therefore, in view of negative liver screen, absence of other risk factors (alcohol), and presence of IR and elevated BMI, both most likely had NASH related cirrhosis. The clinical course was also similar with development of variceal bleeding and HCC.
An earlier report of monozygotic twins with NASH cirrhosis with a similar clinical course10did not involve genetic analysis. Our targeted approach, genotyping candidate genetic markers revealed the presence of effect alleles at 11 of the 14 polymorphic sites studied in these two patients (Table 2). Each of the SNPs investigated in these twins has previously been shown to have an association with indicators of NASH or NAFLD such as hepatic fat content, steatosis and or inflammation,2-7 though none has been directly linked to BMI or IR.These findings are suggestive of a likely cumulative effect of multiple SNPs contributing to the genetic susceptibility that led to the development and progression of NASH. However, we acknowledge that our method could easily have overlooked other polymorphism/ s or novel mutations more directly responsible for the cirrhotic phenotype. Additionally, since the cases were monozygotic twins, their gene profiles were similar- had they been dizygotic, the finding would be more significant.
The PNPLA3 rs738409 polymorphism has been identified as a risk factor for both susceptibility to NAFLD as well as its progression in a number of cohorts.3,4,5,6,11 The encoded lipase mediates lipid hydrolysis in hepatocytes and adipocytes and the G allele (I148M substitution) is associated with increased hepatic fat content3 and steatosis.12 Additionally, carriage of each copy of the G allele confers an additive 2.3 fold risk for HCC.13,14 Both twins were heterozygous for rs738409 and developed HCC. The twins were also homozygous for the rs6006460 SNP in this gene.
The twins were heterozygous for SNPs in apolipoprotein-encoding APOC3 at positions -482 and -455, which are in strong linkage disequilibrium with each other.These variant alleles increasing APOC3 expression have been found to be associated with coronary artery disease, hypertriglycemia, metabolic syndrome15and with NAFLD in Indian males2 in earlier but not in subsequent studies.3-7
A number of recent association studies have reported an association between the GCKR rs7800894 intronic polymorphism and the histological features of NAFLD including hepatic steatosis (odds ratio= 1.45),5 steatohepatitis and fibrosis.6,11,12 Carriage of the minor T allele, associated with decreased insulin release, elevated triglycerides and development of fatty liver,16 and the linked rs1260326 risk allele, were present in the twins in our study (heterozygotes).
Both patients were homozygous for the TRIB1 rs2385114 polymorphism identified through GWAS studies to be associated with elevated ALT, steatosis, steatohepatitis and fibrosis in NAFLD.6 This risk allele was significantly associated with elevated serum triglycerides and cholesterol levels in a GWAS screening for coronary artery disease risk factors.16 Two further polymorphisms, rs1227756 and rs887304, identified by GWAS as associated with lobular inflammation in NAFLD patients,4 were present in both patients (Table 2); they were homozygous for the effect allele of COL13A1 (encoding collagen type XIII α1), and heterozygous for rs887304 in EFCAB4B which encodes a calcium-sensor involved implicated in immune cell activation. Two loci within PPP1R3B (involved in lowering plasma lipids14) were associated with NAFLD in GWAS analyses: rs42406265 and rs11777327.6 However, the risk alleles have not been established.5,11 A recent study described an association between ultrasonographically assessed hepatic steatosis12 with the rs4240624 A allele which was present in the twins (homozygotes).
The FDFT1 (involved in cholesterol biosynthesis) rs2645424 effect allele was not present in our patients. Initial reports of an association of this SNP with histological NAFLD activity score4 were not replicated.
The NCAN Pro92Ser variant had been identified as a NAFLD risk factor in two GWAS studies5,6 and the gene product, neurocan, suggested to have a role in regulating cell adhesion and lipoprotein metabolism. In candidate gene studies, the risk allele was associated with steatosis, inflammation and fibrosis in an American cohort17 but not in obese Asian children with NAFLD.11 Recently, NCAN expression in liver cells was demonstrated and the variant allele, in addition to PNPLA3 rs738409, was associated with the development of HCC (OR=2.29) in patients with alcohol related liver disease.18 The second variant in this chromosomal region, rs58542926 in TM6SF2 was also recently found to be associated with elevated hepatic triglyceride and serum ALT7 and suggested to influence fibrosis progression in NAFLD.19
A twin study (included both concordant and discordant twins), showed that the ADRB2 gene had pleiotropic effects on plasma levels of gammaglutamyltranferase (GGT) and triglycerides, indicating shared genetic codetermination with traits of metabolic syndrome.20 In a more recent twin study, a panel of 10 miRNAs differentiated the twin with NAFLD from the one without.21 MicroRNAs may have an influence beyond genetics, though our twins were concordant and not discordant as in Zarrinpar, et al’s study.21
ConclusionIn conclusion, we describe for the first time, monozygotic twins with NASH related cirrhosis who on genotyping demonstrated heterozygosity for six SNPs (in four gene loci) and homozygosity for five risk-associated alleles in genes that could potentially have been linked to the pathogenesis and progression of NASH. This case series provides further credence to the genetic predisposition to NASH and is suggestive of how the cumulative effect of a number of genes may add up to define a complex trait, i.e. NASH.
Abbreviations- •
ALT: alanine transaminase.
- •
BMI: body mass index.
- •
CAP: controlled attenuation parameter.
- •
GGT: gammaglutamyltransferase.
- •
GWAS: genome-wide association studies.
- •
HCC: hepatocellular carcinoma.
- •
IR: insulin resistance.
- •
LSM: liver stiffness measurement.
- •
miRNA: micro RNA.
- •
NAFLD: non-alcoholic fatty liver disease.
- •
NASH: non-alcoholic steatohepatitis.
- •
PCR: polymerase chain reaction.
- •
SNP: single nucleotide polymorphisms.
- •
TACE: transarterial chemoembolization.
This research was supported by the National Institute for Health Research (NIHR) Nottingham Digestive Diseases Biomedical Research Unit based at Nottingham University Hospitals NHS Trust and University of Nottingham. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health.
Conflict of InterestNone.
Financial SupportNational Institute for Health Research.