Paracoccidioidomycosis is an endemic mycosis caused by members of the Paracoccidioides genus. Brazil remains the focus area and, to a lesser extent, the disease has been reported from Argentina, Colombia and Venezuela.
AimsA Venezuelan Paracoccidioides brasiliensis strain, isolated from a patient diagnosed with chronic multifocal paracoccidioidomycosis, was subjected to whole genome sequencing to provide more insight about Paracoccidioides outside the endemic focus area.
MethodsP. brasiliensis strain CBS 118890 was whole genome sequenced using nanopore; library preparation with the ‘native barcoding genomic DNA kit’ was followed by sequencing on Flongle and MinION flowcells. Batches of strain CBS 118890 were re-identified by sequencing the internal transcribed spacer (ITS) region, and final identification was made based on phylogenetic analysis.
ResultsSurprisingly, the Venezuelan P. brasiliensis strain CBS 118890 turned out to be a Nannizziopsis species. The batches of this strain were ITS sequenced followed by phylogenetic analysis and resulted in the final identification of Nannizziopsis arthrosporioides.
ConclusionsNannizziopsis infections are commonly seen in a wide variety of reptiles, but are particularly rare in human infections. This case underlines the need for molecular characterization of cases that clinically mimic paracoccidioidomycosis but that are serologically negative for Paracoccidioides.
La paracoccidioidomicosis es una micosis endémica causada por especies del género Paracoccidioides. Brasil sigue siendo el área con la mayor incidencia y, en menor medida, se ha informado de casos en Argentina, Colombia y Venezuela.
ObjetivosUna cepa venezolana de Paracoccidioidesbrasiliensis, obtenida de un paciente diagnosticado con paracoccidioidomicosis multifocal crónica, se sometió a secuenciación completa del genoma para obtener más información sobre Paracoccidioides fuera del área de foco endémico.
MétodosSe secuenció el genoma completo de la cepa CBS 118890 de P. brasiliensis mediante la técnica de secuenciación de nanoporos; tras la preparación de la librería con el «native barcoding genomic DNA kit» se procedió a la secuenciación con el Flongle y MinION flowcells. Los lotes de la cepa CBS 118890 se volvieron a identificar mediante la secuenciación de la región del espaciador transcrito interno (ITS), y la identificación final se realizó en función del análisis filogenético.
ResultadosSorprendentemente, la cepa venezolana P. brasiliensis CBS 118890 resultó ser una especie de Nannizziopsis. Los lotes de esta cepa se secuenciaron mediante ITS seguido de un análisis filogenético y dieron como resultado la identificación de la especie Nannizziopsis arthrosporioides.
ConclusionesLas infecciones por Nannizziopsis se observan comúnmente en una amplia variedad de reptiles, pero son particularmente raras en infecciones humanas. Este caso subraya la necesidad de la caracterización molecular de los casos que clínicamente reflejan paracoccidioidomicosis, pero que son serológicamente negativos para Paracoccidioides.
The order Onygenales harbours a large number and wide variety of human fungal pathogens, from dermatophytes causing superficial infections to the life-threatening pathogens that fall under the umbrella of endemic mycoses. The latter includes the genera Blastomyces, Emergomyces, Histoplasma and Paracoccidioides (family Ajellomycetaceae), and Coccidioides (family Onygenaceae).6 Of these fungi, Paracoccidioides is exclusively reported from Latin American countries, being represented by seven species: Paracoccidioides americana, Paracoccidioides brasiliensis, Paracoccidioides restrepiensis and Paracoccidioides venezuelensis (members of the Paracoccidioides brasiliensis species complex), Paracoccidioides loboi, Paracoccidioides lutzii and Paracoccidioides ceti.4,6,17,21 The proposed revision of the Paracoccidioides taxonomy was supported by genome sequencing studies.11,14,20
In 2005, the CBS culture collection, hosted at the Westerdijk Fungal Biodiversity Institute (Utrecht, The Netherlands), received a P. brasiliensis strain from Venezuela that was obtained from a tongue lesion of a patient who was diagnosed with chronic multifocal paracoccidioidomycosis, for which itraconazole (200mg per day) was administered. Unfortunately, given the time that had elapsed since the deposition of the strain, no further clinical information could be obtained. As Paracoccidioides strains from Venezuela were not yet commonly available with whole genome sequence data, we decided to perform long-read nanopore sequencing. For this purpose, the strain was subcultured into 10ml malt extract broth and incubated for two weeks at 28°C. Cells were harvested by centrifugation, followed by genomic DNA extraction as previously described.15 Nanopore library preparation was performed using the ‘native barcoding genomic DNA kit’ (EXP-NBD114 and SQK-LSK109; ONT, Oxford, UK), and the readings were generated with a pre-run – to check the quality of the prepared library – on a Flongle flow cell (FLO-FLG001; ONT), followed by a run onto a MinION flow cell (FLO-MIN106; ONT). Raw data were base called with high-accuracy settings and demultiplexed using Guppy v.6.0.1+652ffd179 (ONT). The FASTQ data from both runs were combined and subsequently subjected to de novo genome assembly using Flye v.2.9-b1768.8 The observed genome size was 24,246,093bp, consisting of 6 contigs representing likely 5 chromosomes plus the mitochondrial genome. The lengths of these 5 chromosomes were 7,230,366bp; 6,417,451bp (=N50); 4,132,340bp; 4,046,396bp, and 2,395,022bp. The circular mitochondrial genome was 24,518bp in length. The coverage of the nuclear genome was 35X and that of the mitochondrial genome 218X. Data were deposited in NCBI Genome under the accession numbers BioProject PRJNA843904, BioSample SAMN28772366, Sequence Read Archive (SRA) SRR19450098 and SRR19450099, and the genome assembly CP098259–CP098264.
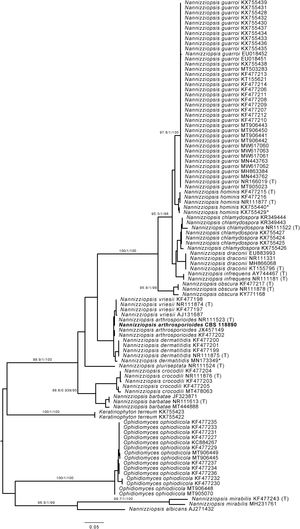
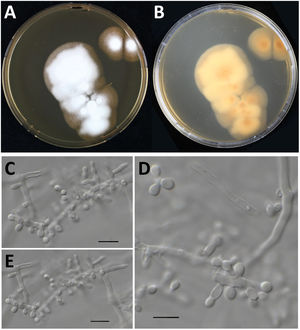
However, when processing the data, it became clear that the sequenced strain CBS 118890 was not P. brasiliensis but the distantly related species Nannizziopsisarthrosporioides (family Onygenaceae), known to behave pathogenic in reptiles.2,18 All available batches of this strain, including the original material stored as oil-preserved slant, were obtained from the CBS culture collection to rule out contamination or erroneous change of material. These strains were all subjected to sequencing the internal transcribed spacer (ITS) region of the ribosomal DNA using primers ITS1 and ITS4, as previously described.5 All batches resembled N. arthrosporioides when ITS sequences were identified against the NCBI nucleotide database (all ITS sequences identical, NCBI GenBank accession number ON725014). We extracted ITS data from the NCBI nucleotide database and performed phylogenetic analyses. Sequences were aligned using MAFFT v7.490, with the strategy setting set to ‘FFT-NS-j’. The phylogenetic tree was built using IQ-tree v1.6.12 with the settings ‘-redo -nt AUTO -nm 1000 -m MFP+MERGE -abayes -alrt 1000 -bb 1000’ and substitution model GTR+F+G4 was automatically calculated by IQ-tree on the basis of Akaike Information Criterion.7,12 In the phylogenetic tree depicted in Fig. 1, strain CBS 118890 clustered tightly together with the other N. arthrosporioides isolates, including the type-strain sequence. Unequivocally, strain CBS 118890 represents N. arthrosporioides, and the original identification as P. brasiliensis was erroneously made based on the clinical presentation. Culture characteristics were also atypical for Paracoccidioides, as can be observed by the colony macro- and micro-characteristics of the CBS 118890 strain in Fig. 2.
Phylogenetic analysis of Nannizziopsis strains. *=Sequences KX755429 and KX755440 were originally deposited as Nannizziopsis guarroi but cluster together with Nannizziopsis hominis. Sequence MN173349 was originally identified by Chen and colleagues (2021) as Nannizziopsisarthrosporioides but is now placed within the group of Nannizziopsisdermatitidis strains.2 (T)=Sequence originated from type-strain of specified species. The scale bar represents the nucleotide substitution ratio. Numbers next to major branches and values >85/0.85/85 represent the support of the likelihood ratio test, Bayesian interference, and bootstrapping, respectively.11Nannizziopsis arthrosporioides strain CBS 118890 is highlighted in bold.
Culture characteristics of Nannizziopsisarthrosporioides CBS 118890. Panels A and B: Culture onto malt extract agar incubated for two-weeks at 35°C, colony is about 5cm in diameter, velvety to powdery (panel A, obverse; panel B, reverse). Panels C–E: Microscopy of a two-week old N. arthrosporioides culture showing aleuriospores and subglobose spores, scale bar=10μm.
At present, there are twelve species recognized within Nannizziopsis: N. arthrosporioides, Nannizziopsis barbatae, Nannizziopsis chlamydospora, Nannizziopsis crocodili, Nannizziopsis dermatitidis, Nannizziopsis draconii, Nannizziopsis guarroi, Nannizziopsis hominis, Nannizziopsis infrequens, Nannizziopsis obscura, Nannizziopsis pluriseptata, and Nannizziopsis vriesii.18,19 The dreaded cause of fungal snake disease, Ophidiomyces ophidiicola, is closely related to the genus Nannizziopsis.9Nannizziopsis infections are commonly seen in a wide variety of reptiles, but are particularly rare in human infections. A handful of human cases caused by N. guarroi, N. hominis, N. infrequens, N. obscura, and undetermined Nannizziopsis species have been reported.1,3,10,13,16,18,19,22 Retrospectively, we can add a human case of N. arthrosporioides infection to this list, an infection that was mimicked as paracoccidioidomycosis. So far, most Nannizziopsis cases in humans have been documented from people living in – or recently immigrated from – Western Africa. The presented case from Venezuela underlines the need for further molecular characterization in cases that clinically reflect paracoccidioidomycosis but that are serologically negative for Paracoccidioides, as they might be Nannizziopsis infections.