Achromobacter spp. are increasingly recognized as emerging pathogens in immunocompromised patients or suffering cystic fibrosis, but unusual in immunocompetent hosts or individuals that underwent surgery. In this study we describe two simultaneous events attributable to two different Achromobacter spp. contaminated sources. One event was related to an episode of pseudo-bacteremia due to sodium citrate blood collection tubes contaminated with Achromobacter insuavis and the other to Achromobacter genogroup 20 infection and colonization caused by an intrinsically contaminated chlorhexidine soap solution. Both threatened the appropriate use of antimicrobials. Molecular approaches were critical to achieving the accurate species identification and to assess the clonal relationship, strengthening the need for dedicated, multidisciplinary and collaborative work of microbiologists, specialists in infectious diseases, epidemiologists and nurses in the control of infections to clarify these epidemiological situations.
Achromobacter spp. son reconocidas con mayor frecuencia como patógenos emergentes en pacientes con fibrosis quística e inmunodeprimidos, pero son inusuales en hospedadores inmunocompetentes o quirúrgicos. En este estudio describimos 2 eventos simultáneos atribuibles a 2 fuentes contaminadas con Achromobacter spp. Uno correspondió a un episodio de seudobacteriemia por tubos de citrato de sodio contaminados con Achromobacter insuavis y el otro a infecciones y colonizaciones debidas al uso de solución jabonosa de clorhexidina intrínsecamente contaminada con Achromobacter genogrupo 20. Ambos episodios pusieron en peligro el uso apropiado de antimicrobianos. Los enfoques moleculares fueron fundamentales para lograr la identificación precisa de las especies y evaluar la relación clonal de los aislamientos, lo que refuerza la necesidad del trabajo perseverante y multidisciplinario de microbiólogos, especialistas en enfermedades infecciosas, epidemiólogos y enfermeras en el control de infecciones para el esclarecimiento de estas situaciones epidemiológicas.
Achromobacter spp. are aerobic, non-fermenting gram negative bacilli that are widely distributed in the environment. Achromobacter xylosoxidans, the first member of this genus, was first described by Yabuuchi and Oyama in 1971, then it was reclassified to the Alcaligenes genus, and later reassigned to the Achromobacter genus. Nowadays Achromobacter comprises 21 species, and some genogroups that have not been fully characterized yet8. Phenotypic species identification, based on biochemical tests, usually renders incorrect or non-conclusive results; for that reason clinical isolates are mostly referred to as A. xylosoxidans. Accurate identification at species level can only be achieved using genotypic methods although these techniques are not used for routine identification12.
Achromobacter spp. have been usually associated with aquatic environments, and frequently colonize aqueous solutions in the hospital settings from where they may infect compromised patients1. Achromobacter spp. are increasingly recognized as important emerging nosocomial pathogens, mainly in immunocompromised hosts, as well as in patients with cystic fibrosis1,13. Although increasingly reported, it is difficult to interpret their clinical significance.
Effective treatment of Achromobacter spp. infections can be challenging due to their natural and acquired multidrug resistance patterns6,7. Moreover, antiseptics and disinfectants are not always able to eliminate the source of Achromobacter spp. in the hospital environments3,10.
Several outbreaks and pseudo-outbreaks caused by Achromobacter spp. have been reported, and many of them have contaminated fluids such as chlorhexidine as the epidemiological “common denominator”2,3,10. Both outbreaks and pseudo-outbreaks are negative as hospital quality indicators, as well as the inappropriate use of antibiotics. Institutional multidisciplinary management is required to prevent infections, outbreaks and pseudo-outbreaks, and the consequent inappropriate use of antibiotics, since antimicrobial stewardship constitutes another indicator of nosocomial quality.
Starting 2013 an increased presence of Achromobacter spp. in blood cultures and clinical samples was observed in a tertiary hospital in Buenos Aires city, leading to this epidemiological study.
Event report 1A descriptive study of Achromobacter spp. positive blood samples, recovered between 2013 and 2016, was carried out retrospectively, in order to characterize a possible outbreak and identify probable sources of contamination.
The incidence of Achromobacter spp. contaminated blood cultures, with respect to the total blood samples, was 51/36286 (0.14%) in 2013, 91/40285 (0.22%) in 2014 and 79/43450 (0.18%) in 2015. True bloodstream infections were confirmed by clinical evaluation and a second positive blood culture5,16. Many of these contaminated samples came from emergency wards (61.5%), and in some cases, other unusual non-fermenting gram negative bacilli were also present.
From June to October 2015, the probable sources of contamination were investigated, including heparin tubes, povidone-iodine solutions, and EDTA and sodium citrate lab tubes. Out of a total of 48 analyzed samples, 4 sodium citrate tubes were positive for Achromobacter spp.
Species identification of the isolates recovered from blood samples (n=6) and blood collection test tubes (n=4) was achieved using genotypic approaches. The nrdA sequences were determined according to Spilker et al., and compared with databases (https://pubmlst.org/achromobacter/)12. The presence of species-specific markers, blaOXA-114, blaOXA-258, blaOXA-364 and blaOXA-243, intrinsic for A. xylosoxidans, Achromobacter ruhlandii, Achromobacter dolens, and Achromobacter insuavis, respectively, were investigated according to Traglia et al. and Papalia et al.11,14. All these bacterial isolates were identified as A. insuavis.
XbaI-pulsed-field gel electrophoresis (PFGE) was performed to compare the isolates recovered from sodium citrate tubes among them, and with those recovered from blood cultures9. A single pulsotype indicative of the presence of the same strain in all samples was observed (Suppl. Fig. S1). Based on these results, it was assumed that these pseudo-bacteremias were originated by the use of contaminated sodium citrate tubes and the subsequent incorrect technique of loading the blood culture bottles, i.e., first inoculating the blood collection test tubes that were contaminated with A. insuavis followed by the inoculation of the blood culture bottles with the same needle generating false positive blood cultures. In order to avoid these pseudo-outbreaks, personnel retraining regarding the correct sample loading technique was conducted and institutional guidelines were designed ad hoc.
Event report 2A strikingly high number of Achromobacter spp. isolates in clinical specimens other than blood samples was also detected in our hospital, which led to an epidemiological study to characterize a possible outbreak and to identify a common denominator.
A total of 94 Achromobacter spp. positive samples, recovered from 59 patients from March 2014 to June 2016, were included in this study (Suppl. Fig. 2).
Each of these episodes was classified as Achromobacter spp. infection or colonization after reviewing the patients’ clinical histories. Patients with a positive culture and symptoms compatible with infection without any other apparent focus and with specific antibiotic treatment selected for these microorganisms were categorized as infected by Achromobacter spp., while patients with a positive culture for Achromobacter spp. without any clinical symptoms compatible with infection were categorized as colonized (Table 1). Twenty eight percent (28%) of the analyzed cases were interpreted as infections and the remaining 72% as Achromobacter spp. colonization. Mortality rate after 30 days was 1%, although it was unrelated to the presence of Achromobacter spp. in cultures. Ninety five percent (95%) of these cultures were recovered from patients that either were hospitalized for 48h, or with previous hospitalization (less than 90 days), or that underwent surgery, thus categorized as hospital-acquired infections/colonizations. Clinical specimens are shown in Table 2.
Characteristics of patients with microbiological samples (not blood cultures) positive for Achromobacter spp.
| Patient information (n: 59) | Number of patients |
|---|---|
| Age | 49 years (average) |
| Sex | 21 female/38 male |
| Underlying disease | |
| Immunocompromised patients | 16 |
| Solid organ and bone marrow transplantation | 3 |
| Solid tumor surgery | 12 (6 bone tumors) |
| Others | 1 |
| Other comorbidities | |
| Diabetes | 6 |
| Hepatic failure | 2 |
| Peripheral cardiovascular disease | 2 |
| Neurological disease | 4 |
| Chronic respiratory disease | 4 |
| Orthopedics and traumatology patients | 38 (6 bone tumors) |
| In-patients in internal medicine | 7 |
| In-patients in critical units | 10 |
| Surgery patients | 4 |
| Achromobacter spp. infection | 16 |
| Achromobacter spp. colonization | 43 |
| 30-Day mortality | 1 (unrelated death) |
Distribution of Achromobacter spp. positive clinical specimens.
| Clinical specimens (n=94) | 2014 (n=13) | 2015 (n=54) | 2016 (n=27) |
|---|---|---|---|
| Respiratory samples: tracheal aspirate, bronchoalveolar lavage, sputum | 1 | 7 | 5 |
| Post-orthopedic surgery samples: bone, soft tissue, wound | 10 | 39 | 15 |
| Other samples: breast, wound, gastric fluid, duodenum, catheter tip, urine, skin, hematoma | 2 | 8 | 7 |
Among Achromobacter spp. positive samples, a broad dominance was observed in those recovered from trauma surgeries and surgeries with anfractuous open wounds, performed at the orthopedic operating room (64/94). Twenty nine percent (29%) of the patients were assisted in clinical areas and 59% in critical care areas; in 23% previous washing with soapy chlorhexidine was recorded. Likewise, Achromobacter spp. were also recovered from other clinical samples, and other areas of the institution, suggesting a common source of extensive use throughout the hospital.
In April 2016, environmental, instrumental, antiseptic and disinfectant samples from 5 different operating rooms were analyzed. Achromobacter spp. was identified in 15/17 samples of 4% soapy chlorhexidine, recovered from all operating rooms, as well as in unopened containers stored in this center since 2014, indicating intrinsic contamination. Soapy chlorhexidine samples were removed from the spray bottle with pipettes to avoid touching the container. The soapy chlorhexidine samples were cultured in a PF Bact-Alert® flask (bioMérieux). Incubation was carried out for 5 days to rule out the possibility of a negative result. Prior to inoculation, a two-fold dilution of the sample was performed using sterile human plasma. The final inoculation volume was 5ml (2.5ml of antiseptic solution and 2.5ml of human plasma).
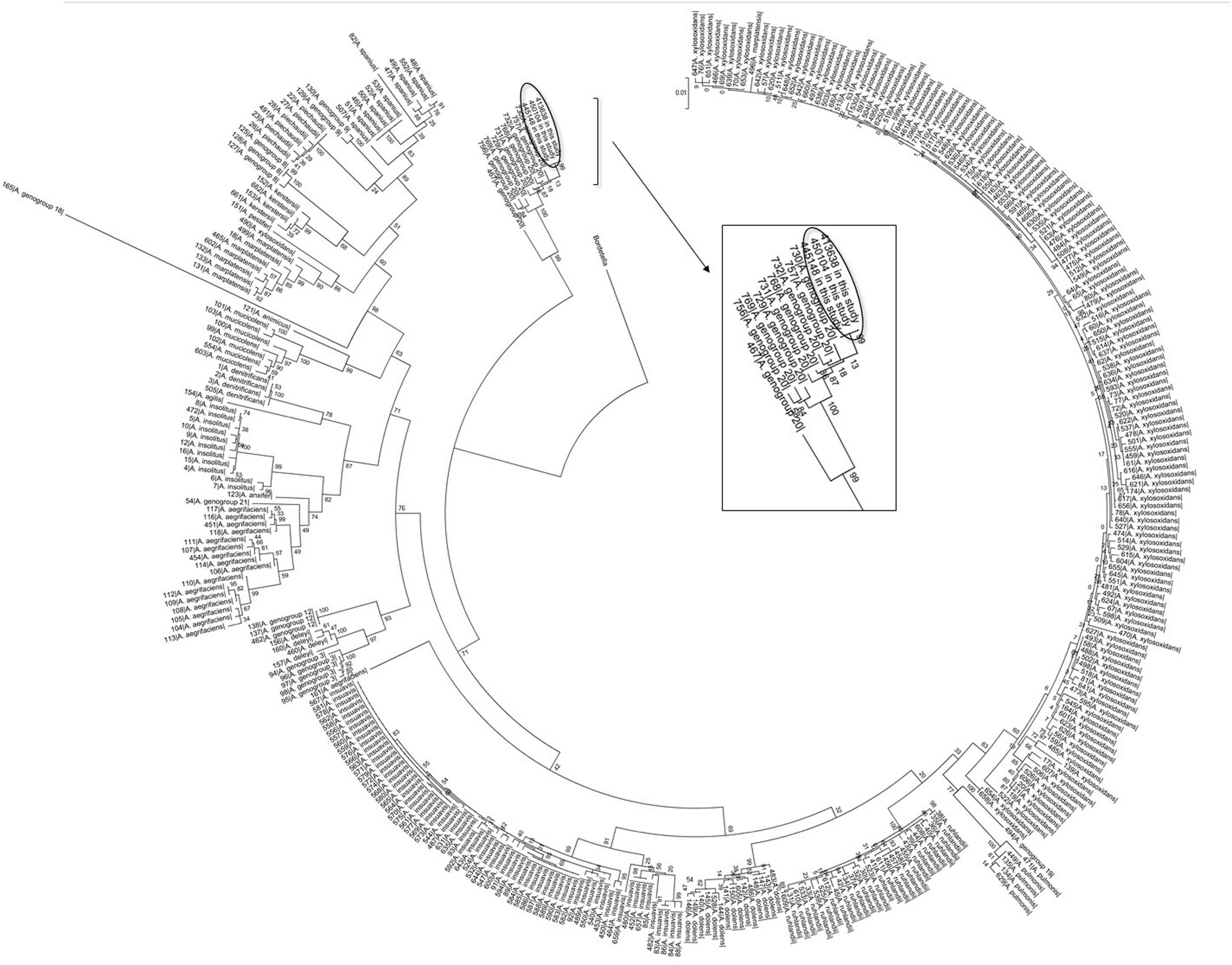
Twenty-five (25) isolates recovered from clinical specimens and from the chlorhexidine soap solution were identified by nrdA sequencing12, corresponding to A. xylosoxidans (n: 1) and Achromobacter spp. (n: 24). A multilocus sequence-typing (MLST) scheme was conducted to identify those isolates which could not be unambiguously identified by nrdA sequencing. For this purpose, amplification and sequencing of inner fragments of seven housekeeping genes were conducted. Concatenate sequences (2254 nucleotides) of these seven fragments were obtained for those isolates included in this study and for 362 Achromobacter spp. available in the database (Suppl. Table 1). An alignment of all concatenates was performed using the program ClustalX 2.1. The statistical selection of best-fit model of nucleotide substitution was assessed by JModelTest. Maximum Likelihood estimation was conducted using MEGA 5.05. Bootstraps with 1000 re-sample matrices were executed to assess the statistical support for the identified groups. The dendrogram obtained with Maximum Likelihood is depicted in Figure 1, showing that these isolates grouped with Achromobacter genogroup 20. However, they could correspond to a new species of the genus Achromobacter (a long-term analysis out of the scope for the present study).
XbaI-pulsed-field gel electrophoresis (PFGE) was performed to compare the Achromobacter genogroup 20 isolates, recovered both from clinical specimens and from the chlorhexidine soap solution; PFGE analysis showed 4 different pulsotypes (Suppl. Fig. 3).
Surgical wound lavage with chlorhexidine soap solution was interpreted as possibly responsible for the high colonization/infection rate in trauma surgeries, although this practice could only be confirmed in a low percentage of patients by surgery records. It should be mentioned that surgical wound lavage with sterile physiological solution is recommended in this institution.
The intrinsic contamination of the antiseptic soapy chlorhexidine solution prompted the following measures: removal of all 4% chlorhexidine soap solution bottles from the entire facility; notification to the supplier about the presence of Achromobacter spp. in unopened bottles of 4% chlorhexidine soap solution; and the complaint to the relevant health authorities, the National Administration for Drugs, Food and Medical Technology (Administración Nacional de Medicamentos, Alimentos y Tecnología Médica (ANMAT)).
Since this antiseptic was removed, between May and June 2016, the presence of Achromobacter spp. was detected only once in the course of the next 18 months, corresponding to in a clinical sample from the orthopedic operating room in a post-intervention toilet, which was interpreted as contamination. The withdrawal of all the vials in the institution continued to be reinforced.
DiscussionIn this study we describe two simultaneous events attributable to two different Achromobacter spp. contaminated sources, in addition to incorrect procedures that facilitated their dissemination. One event was related to an episode of pseudo-bacteremia due to sodium citrate blood collection tubes contaminated with A. insuavis added to a deficient technique in the loading of blood culture bottles, which caused their contamination. The other corresponded to Achromobacter genogroup 20 infections and colonizations caused by an intrinsically contaminated chlorhexidine soap solution, which was probably used in surgical wound healing and other clinical scenarios.
The undesirable effect of contaminated blood culture and their financial impact is well known, being independently associated with increased laboratory charges, hospitalization admission and inappropriate use of intravenous antibiotics4.
The risk of antiseptic solution contamination, especially by gram negative bacilli, is well documented in the medical literature15. Moreover, pseudo-bacteremia outbreaks have been already associated with Achromobacter spp. contaminated chlorhexidine aqueous solutions3,10. However, outbreaks with a predominance of clinical samples related to surgical wounds, such as the one described in this study, are infrequent. In other Achromobacter spp. pseudo-outbreaks, contamination of the chlorhexidine spray pipette has been described, although it was ruled out in this study by direct sampling of chlorhexidine with sterile pipettes avoiding the passage through the spray pipette of the container.
The investigation process of these events was cumbersome, in part due to the presence of two different species of the same genus that could not be identified by phenotypic tests in the routine laboratory, added to the great variety of sample types and different hospital areas involved. Molecular approaches were critical to achieving the accurate species identification and to assess the clonal relationship of the isolates. The importance of the multidisciplinary work of microbiologists, infectious disease specialists, epidemiologists and nurses in infection control should be emphasized, with great perseverance in elucidating this epidemiological situation.
This study confirms, once more, the likelihood of aqueous chlorhexidine solution contamination by ubiquitous environmental hydrophilic microorganisms such as Achromobacter spp. with the hazard of related hospital outbreaks and pseudo-outbreaks. In addition, it highlights the importance of conducting outbreak investigations, persisting even with initial disappointing findings, implementing molecular studies, alerting against this type of deviations, and highlighting the need to reinforce microbiological controls (including long-term microbiological stability) in antiseptic production companies.
FundingThis work was partially supported by grants from UBACyT to M. Radice (20020150100174BA) and G. Gutkind (20020130100432BA) and by ANPCYTPICT-2017-3996 to Mariana Papalia.
M. Radice, G. Gutkind, M. Papalia are members of Carrera del Investigador Científico (CONICET).
Hospital Italiano – The funds correspond to the budget assigned to the infection committee.
Conflict of interestNone declared.