There is limited information about the prevalence and antimicrobial susceptibility of coagulase-positive Staphylococcus (CoPS) strains in veterinary settings in Chile. The aim of this observational study was to identify and characterize CoPS strains from dogs, owners, veterinary professionals and surfaces in a veterinary teaching hospital at Universidad de Chile to determine the presence of methicillin-resistant strains and evaluate the genetic relationship among the strains. Veterinarians (n=24), surfaces (n=10), and healthy dogs (n=40) and their respective owners (n=40) were sampled for CoPS. Isolates were identified by PCR and antimicrobial susceptibility was assessed by the disk diffusion method and MIC. The presence of the mecA gene was evaluated by PCR, and the genetic relationship among the strains was established by PFGE. A total of 45 CoPS strains were obtained, eight from veterinary professionals, three from hospital surfaces, eight from owners and 26 from dogs. Nine of the strains were resistant to methicillin (20%), and all of them carried the mecA gene. A high percentage of the strains was resistant to clindamycin (33.3%). Additionally, the isolated CoPS showed high genetic diversity. This study suggests that veterinarians are in high risk of harboring methicillin-resistant CoPS (25% versus 2.5% from owners) and our results provide evidence that clindamycin could not be an empiric alternative for CoPS in the analyzed hospital. This is the first report of methicillin-resistant CoPS in veterinary settings in Chile, considering humans, pets and surfaces.
Existe información limitada sobre prevalencia y sensibilidad antimicrobiana de cepas de Staphylococcus coagulasa-positivas (CoPS) en entornos veterinarios en Chile. El objetivo de este estudio observacional fue identificar y caracterizar cepas CoPS de perros, dueños, veterinarios y superficies de un hospital veterinario de la Universidad de Chile, determinar la presencia de cepas meticilino-resistentes y evaluar la relación genética entre las cepas. Se colectaron muestras de veterinarios (n=24), de superficies hospitalarias (n=10) y de perros sanos (n=40) y sus respectivos dueños (n=40). Los aislamientos se identificaron mediante PCR y la sensibilidad antimicrobiana se evaluó por difusión en discos y CIM. También se empleó PCR para detectar la presencia del gen mecA; la relación genética entre las cepas se estableció mediante electroforesis de campos pulsantes (PFGE). Se obtuvo un total de 45 cepas de CoPS, 8 de veterinarios, 3 de superficies hospitalarias, 8 de dueños y 26 de perros. Nueve cepas fueron meticilino-resistentes (20%), todas portadoras del gen mecA. Un porcentaje importante de cepas fue resistente a clindamicina (33,3%). Además, las cepas aisladas mostraron una alta diversidad genética. Este estudio sugiere que los veterinarios tienen alto riesgo de portar CoPS resistentes a meticilina (25% versus 2,5% propietarios). Asimismo, nuestros resultados proporcionan evidencia de que la clindamicina podría no ser una alternativa empírica para CoPS en el hospital analizado. Este es el primer estudio de CoPS meticilino-resistentes en entornos veterinarios en Chile que considera humanos, mascotas y superficies.
The skin of mammals is colonized by a large number of microorganisms that are collectively known as normal microbiota, which includes several species of the genus Staphylococcus47. They are normally considered harmless, but they can be a threat to the health of their hosts by acting as opportunistic pathogens and being reservoirs of antibiotic resistance genes, increasing the potential for resistance to antibiotic therapy38. The staphylococci with the highest clinical relevance are coagulase producers37, mainly S. aureus and those from the intermedius group, particularly S. pseudintermedius50. Despite the fact that S. pseudintermedius rarely causes illness in humans, with reports ranging from soft tissue infections to bacteremia, its prevalence may be underestimated, since S. pseudintermedius may be erroneously identified as S. aureus due to its similar biochemical characteristics7,48. Establishing the difference between species is particularly relevant when evaluating behavior against antimicrobials, since the cut-off points to define susceptibility and resistance to methicillin vary according to the species7, unfortunately the characterization of species is not a routine practice in our region.
In general, it can be established that most staphylococcal infections can be easily controlled with antibiotics; however, in recent years different strains of Staphylococcus spp. have proved to be resistant to the most commonly used antibiotics such as macrolides, lincosamides, tetracyclines, gentamicin, cephalosporins and other β-lactams27. The most relevant resistance in Staphylococcus is the mecA-mediated methicillin resistance, which is globally spread, turning ineffective the most widely used and efficient class of antimicrobials to treat staphylococcal infections1. This gene is harbored in a mobile genetic element known as Staphylococcal Chromosomal Cassette (SCCmec), and encodes an altered penicillin-binding protein (PBP) with low affinity for β-lactams1. The emergence and spread of methicillin-resistant Staphylococcus (MRS) in both hospital and community settings pose a major threat to global health34. In general, the prevalence of methicillin-resistant strains has increased rapidly over time, leading to major human and animal health problems, especially in hospital settings10. It is well-known that methicillin-resistant strains have higher levels of resistance against various classes of antibiotics compared to methicillin-susceptible strains16,43,45, and the direct cost to treat methicillin-resistant Staphylococcus aureus (MRSA) infections versus methicillin-susceptible S. aureus in humans is 1.5 to 3 times higher3.
To date, there are no antimicrobial resistance stewardship programs for any animal species, as well as no published studies of antimicrobial use in companion animals in Chile13; currently available information regarding antibiotic resistance in South America is mainly focused on human health and studies on determining the prevalence of MRS strains in companion animals in South America are scarce. Among these studies, there is a study carried out in Brazil reporting the presence of MRSA in a cat and a methicillin-resistant S. pseudintermedius (MRSP) in a dog35 and another one from Argentina informing that MRSA prevalence was much higher than in other studies in the United States where prevalence estimates varied from 0% to 6%10,39 should be highlighted. Moreover, a study carried out in Chile describing 11 strains of coagulase-positive Staphylococcus (CoPS) carrying the mecA gene in cats14 and two studies in Argentina that characterized 1012 and three46 strains of MRSP isolated from canine clinical specimens. Another study in Chile analyzed CoPS strains isolated from dogs with pyoderma and external otitis, not finding any MRS strains32. Despite the high proximity of pets and humans, the importance of companion animals as reservoirs of human infections is still poorly understood27.
The aim of the present study was to identify and characterize CoPS strains from dogs, owners, veterinary professionals and the environment in a veterinary teaching hospital in Chile, to determine the presence of methicillin-resistant strains and to evaluate the genetic relationship between the CoPS strains.
Materials and methodsRegulatory approvalsThe study was approved by the Ethical Committee of Facultad de Ciencias Veterinarias y Pecuarias, Universidad de Chile, authorization code No. 07-2016. An informed consent of the owners and veterinarians was obtained prior to sampling.
SamplingThe sampling was conducted during the April-August period, 2016, and samples were obtained from dogs (n=40) and owners (n=40), veterinarians (n=24), and surfaces (n=10) from a teaching veterinary hospital at the Universidad de Chile. In the case of dogs, healthy patients, of any breed, age and sex, taken to the hospital for vaccinations or elective surgical procedures were sampled. They were clinically evaluated by a veterinarian at the hospital, who confirmed the patient's health status at the sampling time. The use of antibiotics and/or corticosteroids 15 days before taking samples was considered an exclusion factor, for both dogs and humans.
Prior to sampling, the owner signed an informed consent developed under the recommendations of the World Health Organization (supplementary material). Owners and veterinarians were taught to take their own nasal swab sample as previously described33 and a sample was taken from the canines’ perianal area and their nasal mucosa. All samples were stored at 4°C for up to 6h before processing. Regarding the surfaces, five hospitalization cages, the surgical tables of the two surgical wards, a care table in the procedure room, and the care table of two hospital consultation rooms were sampled. All samples were taken with a sterile swab in amies transport medium (Copan, California, USA). A quadrant of 4cm2 was selected in which the swab was slipped for 30s covering the entire area. The samples were collected 3h after cleaning with quaternary ammonium and were taken as previously described11. The samples were stored at 4°C for up to 6h until they were processed.
All samples were cultured in mannitol salt agar plates (MSA, Becton Dickinson GmbH Heidelberg, Germany) for 24h at 37°C. From each plate, a maximum of three colonies with morphology compatible with Staphylococcus spp. was selected to perform the catalase test, coagulase test and Gram staining, as previously described32; but only one isolate from each sample was considered for analysis. The catalase and coagulase positive strains, and confirmed Gram-positive round shape bacteria, were stored in a 2:1 mixture of trypticase broth and 50% glycerol (v/v), at −80°C, for subsequent species characterization.
Molecular identification of the Staphylococcus speciesIn order to identify species of Staphylococcus, a simple PCR protocol was carried out with primers previously described by Sasaki et al.41 DNA was extracted from 5 fresh colonies grown on MSA, by boiling at 100°C for 15min, and suspended in 500μl of sterile distilled water, DNA concentration and purification were evaluated by spectrophotometry prior to PCR. Subsequently, the suspension was centrifuged at 14000×g for 5min. The amplification program used consisted of an initial denaturation step of 2min at 98°C, 30 cycles with 30s at 95°C (for denaturation), 57°C for 30s and 72°C for 30 s (for annealing and polymerization) to end with a final extension of 2min at 72°C. The products were separated on a 1.5% agarose gel in 1× tris–acetate–EDTA (TAE) buffer (Thermo Scientific™, Vilnius, Lithuania) and taken to a 1× TAE buffer electrophoresis chamber at 90V for 60min. DNA visualization after electrophoresis was performed using a UV transilluminator (UVP transilluminator M-20V, Jena, Germany), after staining the gels for 40min in ethidium bromide (0.5mg/ml). PCR reactions were carried out in a Multigene Gradient thermocycler (Labnet International Inc, New Jersey, USA) using 15μl of master mix (GoTaq Green Master Mix, Promega, Madison, USA), forward and reverse primer at a concentration of 1μM and 5μl of DNA template. The S. aureus ATCC 25923 and S. pseudintermedius ATCC 49444 strains were used as reference strains. A list of primers used in this study is shown in Table 1.
List of the primers used for species identification and detection of the mecA gene.
| Target species or gene | Primer | Sequence (5′→3′) | Molecular weight (bp) | Reference |
|---|---|---|---|---|
| S. aureus | au-F3 | TCGCTTGCTATGATTGTGG | 359 | Sasaki et al. (2010) |
| au-nucR | GCCAATGTTCTACCATAGC | |||
| S. pseudintermedius | pse-F2 | TGGGCAGTAGGATTCGTA | 926 | Sasaki et al. (2010) |
| pse-R5 | CTTTTGTGCTTCCTTTTGG | |||
| mecA | mecA1 | TGTCCGTAACCTGAATCAGC | 519 | Ishihara et al. (2010) |
| mecA2 | TGCTATCCACCCTCAAACAG |
Phenotypic resistance was determined for all isolated CoPS strains using the agar diffusion method, according to the recommendations of the Clinical and Laboratory Standards Institute documents M100-S-249 and VET01-S28. The following panel of antimicrobials was used: oxacillin (1μg), cefoxitin (30μg), amoxicillin/clavulanic acid (20/10μg), gentamicin (10μg), amikacin (30μg), tetracycline (30μg), doxycycline (30μg), enrofloxacin (5μg), ciprofloxacin (5μg), clindamycin (2μg) and erythromycin (15μg) (Oxoid, Hampshire, United Kingdom). S. aureus ATCC 25923 strain was used as a reference. Methicillin-resistance was evaluated following the CLSI recommendations, considering the different cut off points of the different species, and the oxacillin disk for S. pseudintermedius and cefoxitin disk for S. aureus8,9. Additionally, and in order to quantify the level of resistance, the MIC was also determined according to the method standardized by the CLSI8,9. The following antimicrobials were tested: oxacillin, cefoxitin, enrofloxacin, clindamycin, and vancomycin. S. aureus ATCC 29213 was used as the reference strain.
Detection of the mecA geneThe presence of the mecA gene was determined in all the strains through the amplification of an intragenic fragment of this gene by PCR, yielding a product of 519bp. S. aureus ATCC 43300 was used as the reference strain. Subsequently, the same program and method to identify the species was used. The primers used were those described by Ishihara et al.20, and are listed in Table 1.
Pulsed field gel electrophoresis (PFGE) typing of the CoPS species:The genetic relationship among the isolated CoPS was conducted using a macrorestriction assay of the total DNA of each strain with the SmaI enzyme and the restriction fragments were separated by PFGE at Laboratorio de Investigación en Agentes Antibacterianos of the Universidad de Concepción. For the genotyping of the strains, we used the PFGE methodology described by the Instituto de Salud Pública de Chile which is based on the protocol standardized by CDC Canada for PFGE of SAMR28. The resulting agarose gel was visualized and photographed in the UVIdoc HD5 photodocumentator (Uvitec, England, United Kingdom). Fingerprint analysis was performed using the BioNumerics® software v.6.611 (Applied Maths). The resulting dendrogram was constructed using the unweighted pair group method for arithmetic averages and the Dice band-based similarity coefficient. Isolates were defined as epidemiologically related (same cluster) if they shared ≥95% similarity on the dendrogram.
Statistical analysisThe Chi-square test was used to seek differences in the isolated percentages between veterinarians and owners
ResultsStrain identificationA total of 45 CoPS strains were obtained: Eight CoPS strains were obtained from the 24 veterinarians sampled, three strains from 10 hospital surface samples, eight strains from the nasal mucosa of the 40 owners, and 26 from the canines (10 from the nasal mucosa and 16 from the perianal area).
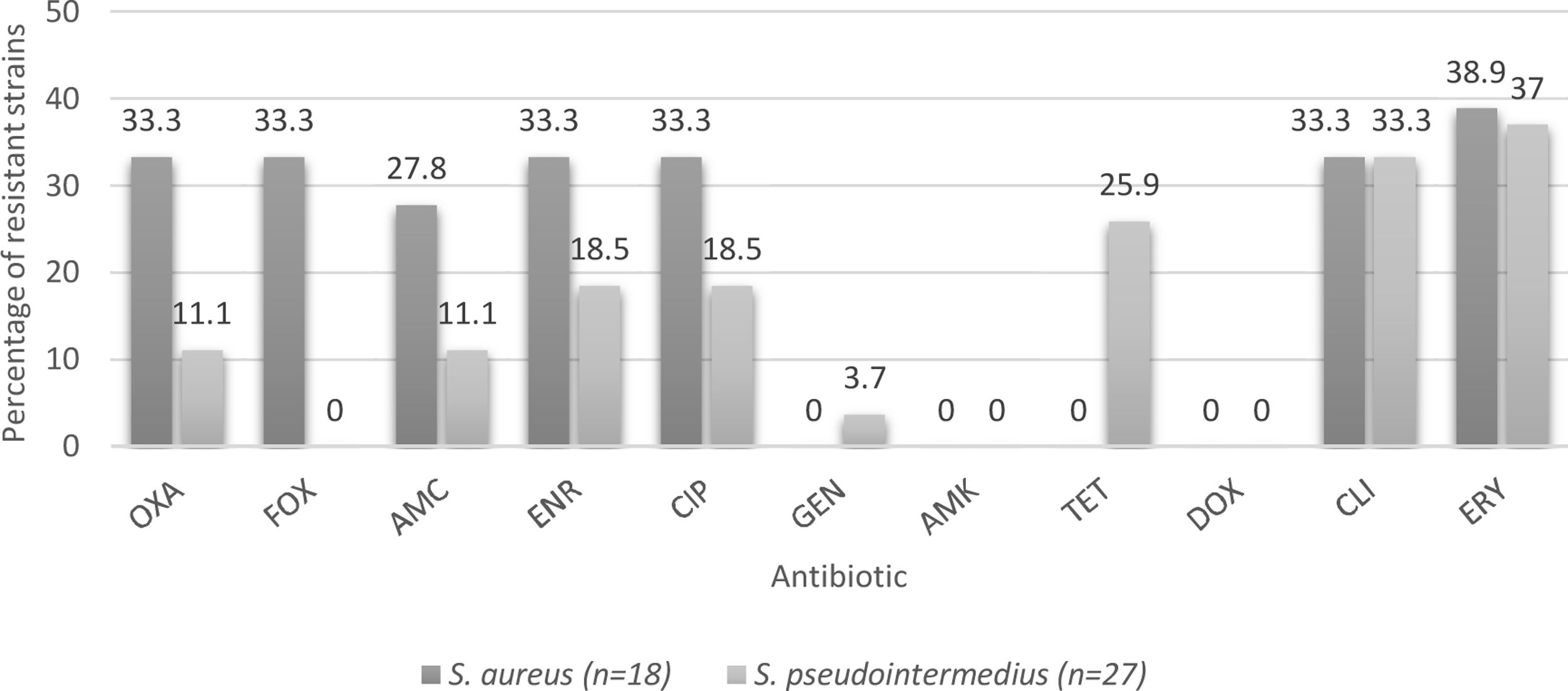
Of the 45 strains identified, 18 were S. aureus and 27 were S. pseudintermedius. In the case of the veterinarians, seven were S. aureus and one S. pseudintermedius. The three strains isolated from hospital surfaces were identified as S. pseudintermedius; while seven strains from the owners were S. aureus and one S. pseudintermedius. Twenty-two strains isolated from the canines were S. pseudintermedius and 4 were identified as S. aureus.
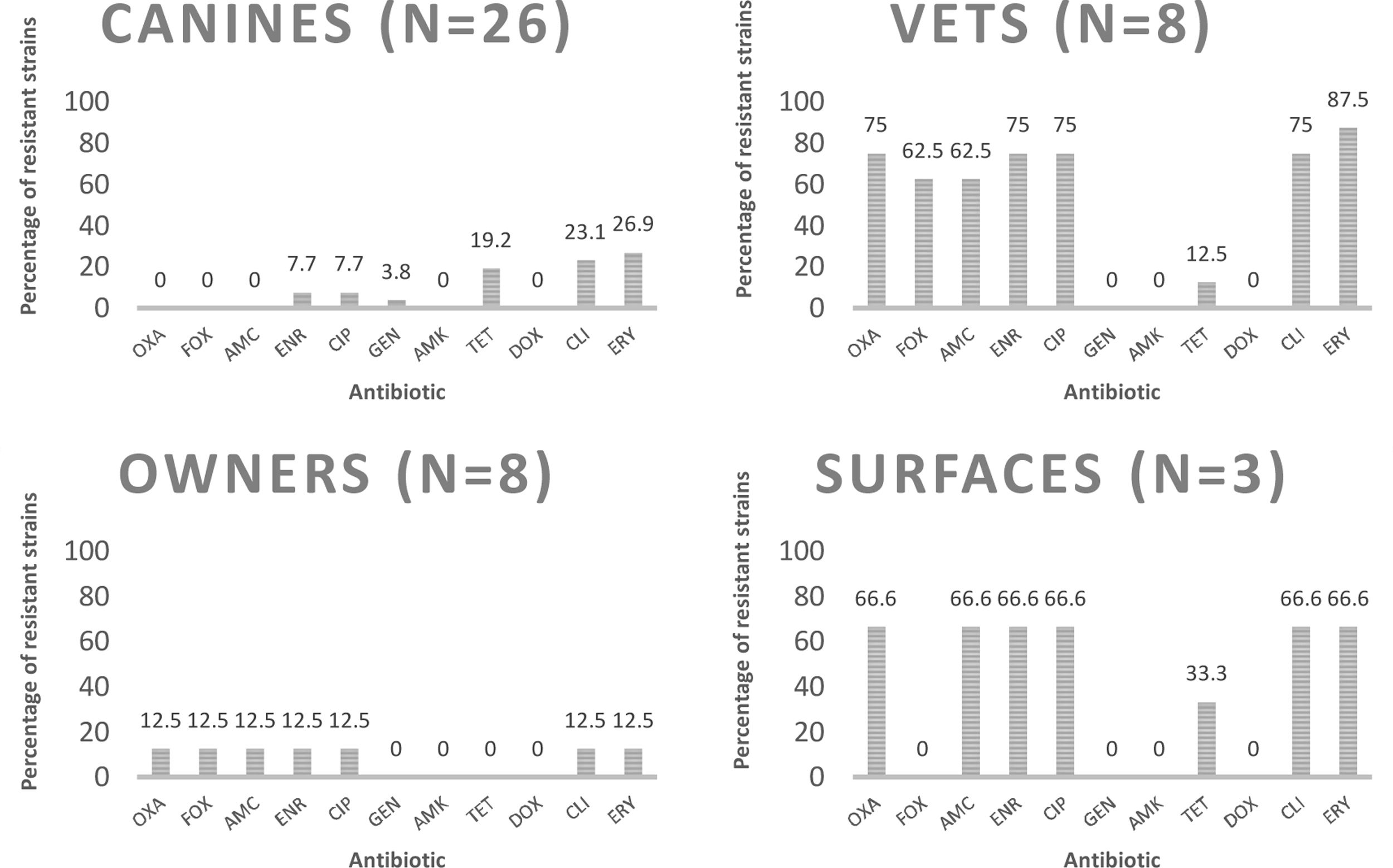
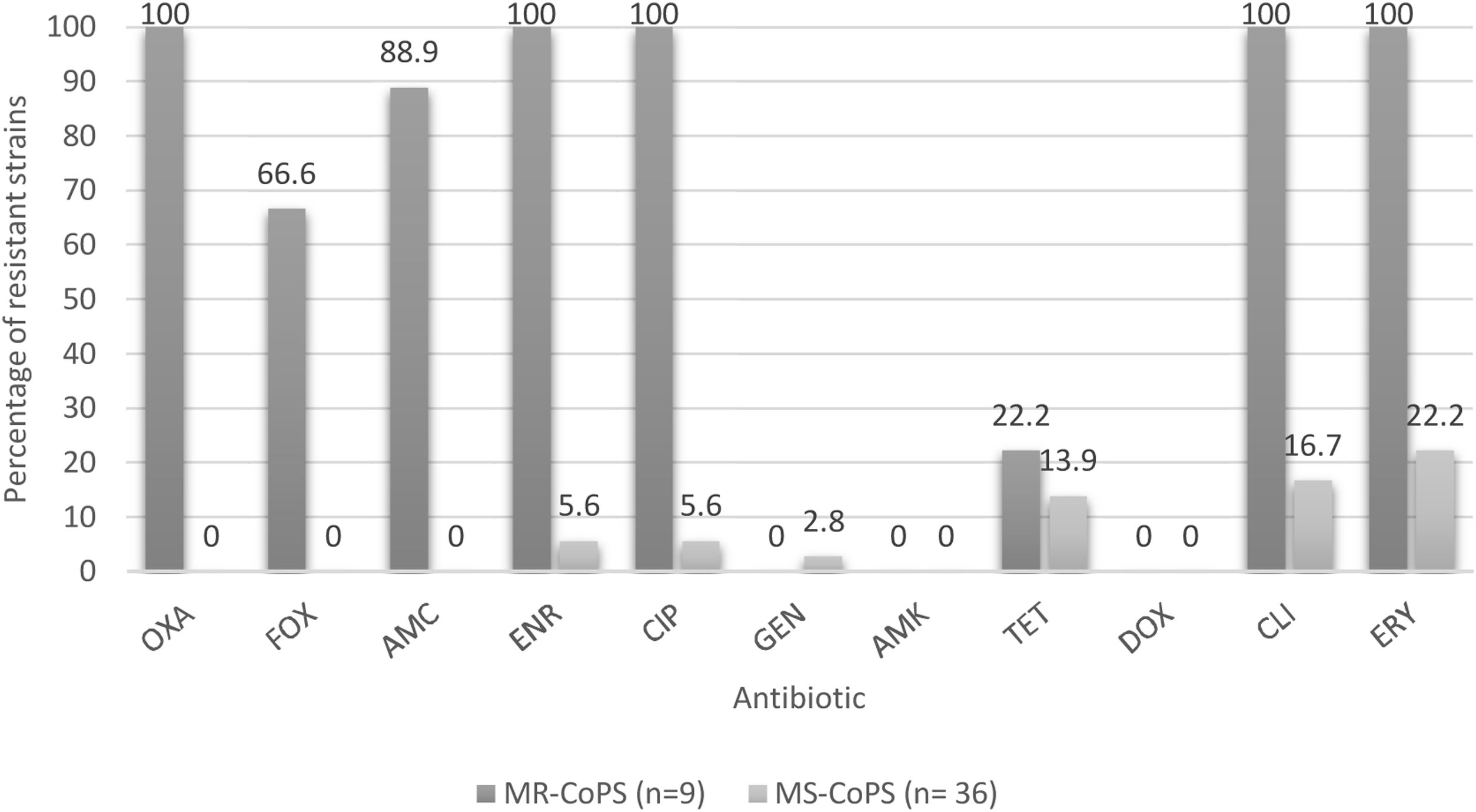
Determination of phenotypic resistance to antibioticsEight (17.8%) of the analyzed strains were susceptible to all tested antimicrobials, and 15 (33.3%) were characterized as multidrug-resistant isolates (resistant to three or more different groups of antibiotics). Percentages of resistant strains considering both species were 37.8% to erythromycin; 33.3% to clindamycin; 24.4% to enrofloxacin and ciprofloxacin; 17.8% to amoxicillin/clavulanic acid; 15.6% to tetracycline; 13.3% to cefoxitin; and 2.2% to gentamicin. Nine strains (20%) were resistant to oxacillin; six of them were obtained from veterinarians; two from hospitalization cages and one from an owner. The characterization of methicillin-resistant strains is shown in Table 2. The percentages of resistance according to the source, species and methicillin susceptibility are listed in Figures 1–3 respectively.
Characteristic of methicillin-resistant strains isolates from veterinary settings in Chile.
| ID | Source | Species | mecA | Resistant profile | Oxacillin | Cefoxitin | ||
|---|---|---|---|---|---|---|---|---|
| KB (mm) | CIM (μg/ml) | KB (mm) | CIM (μg/ml) | |||||
| MV 1 | V | S. aureus | Yes | OXA ENR CIP ERI FOX AMC CLI | 0/R | 64/R | 16/R | 16/R |
| MV 2 | V | S. aureus | Yes | OXA ENR CIP ERI FOX CLI | 0/R | 64/R | 16/R | 32/R |
| MV 3 | V | S. pseudintermedius | Yes | OXA ENR CIP ERI DOX AMC CLI TET | 0/R | >64/R | 26/S | 4/S |
| MV 9 | V | S. aureus | Yes | OXA ENR CIP ERI FOX AMC CLI | 0/R | 64/R | 16/R | 32/R |
| MV 18 | V | S. aureus | Yes | OXA ENR CIP ERI FOX AMC CLI | 0/R | 64/R | 15/R | 32/R |
| MV 21 | V | S. aureus | Yes | OXA ENR CIP ERI FOX AMC CLI | 0/R | 32/R | 16/R | 16/R |
| Equi 9 | S | S. pseudintermedius | Yes | OXA ENR CIP ERI AMC CLI | 0/R | >64/R | 28/S | 4/S |
| Equi 10 | S | S. pseudintermedius | Yes | OXA ENR CIP ERI DOX AMC CLI TET | 0/R | >64/R | 26/S | 4/S |
| Prop 36 | O | S. aureus | Yes | OXA ENR CIP ERI FOX AMC CLI | 0/R | 32/R | 15/R | 16/R |
V: Veterinarian; S: surfaces; O: owner; OXA: oxacillin; ENR: enrofloxacin; CIP: ciprofloxacin; ERI: erytromicin; FOX: cefoxitin; AMC: amoxicillin/cavulanic acid; CLI: clyndamicin; KB: Kirby–Bauer; CIM: Minimal Inhibitory Concentration; R: resistant; S: susceptible.
Percentages of resistance of CoPS strains isolated from a teaching veterinary hospital to the different antimicrobials tested according the source. OXA: oxacillin; FOX: cefoxitin; AMC: amoxicillin/clavulanic acid; ENR: enrofloxacin; CIP: ciprofloxacin; GEN: gentamicin; AMK: amikacin; TET: tetracycline; DOX: doxycycline; CLI: clindamycin; ERY: erythromycin.
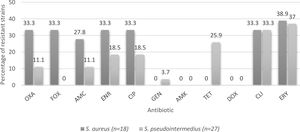
Percentages of resistance of CoPS isolated from a teaching veterinary hospital according the species of Staphylococcus. OXA: oxacillin; FOX: cefoxitin; AMC: amoxicillin/clavulanic acid; ENR: enrofloxacin; CIP: ciprofloxacin; GEN: gentamicin; AMK: Amikacin; TET: tetracycline; DOX: doxycycline; CLI: clindamycin; ERY: erythromycin.
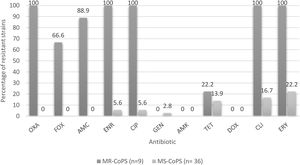
Percentages of resistance of CoPS isolated from a teaching veterinary hospital according methicillin susceptibility. OXA: oxacillin; FOX: cefoxitin; AMC: amoxicillin/clavulanic acid; ENR: enrofloxacin; CIP: ciprofloxacin; GEN: gentamicin; AMK: Amikacina; TET: tetracycline; DOX: doxycycline; CLI: clindamycin; ERY: erythromycin; MR-CoPS: Methicillin-resistance coagulase-positive Staphylococcus; MS-CoPS: Methicillin-sensitive coagulase-positive Staphylococcus.
Eleven strains were resistant to oxacillin according to their MIC (≥4μg/ml for S. aureus and 0.5μg/ml for S. pseudintermedius), six of them corresponded to strains isolated from the veterinarians (five S. aureus, one S. pseudintermedius), two from hospitalization cages (S. pseudintermedius), one from an owner (S. aureus) and two from the canines; these two strains from canines were S. pseudintermedius-negative for the presence of the mecA gene.
Eight strains were resistant to cefoxitin according to the MIC (≥8μg/ml), five of them were isolated from the veterinarians (S. aureus), two from the owners (S. aureus) and one from a canine (S. pseudintermedius). Regarding enrofloxacin, 14 strains were resistant (MIC≥4μg/ml); six of them were isolated from the veterinarians (five S. aureus and one S. pseudintermedius), two from hospitalization cages (S. pseudintermedius), two from the owners (S. aureus) and four from the canines (S. pseudintermedius). Twenty one (21) strains were resistant to clindamycin (MIC≥4μg/ml); six of them were isolated from the veterinarians (five S. aureus and one S. pseudintermedius), two from hospitalization cages (S. pseudintermedius), three from the owners (S. aureus), and 10 from the canines (S. pseudintermedius); while two strains isolated from a veterinarian (S. aureus) and a canine (S. pseudintermedius) exhibited intermediate sensitivity (MIC 1–2μg/ml). Finally, two strains were resistant to vancomycin (MIC≥32μg/ml), both isolated from the owners (S. aureus), while four strains exhibited intermediate susceptibility (MIC 4–8μg/ml), two from the surfaces (S. pseudintermedius) and two from the canines (one of each species). MIC50 and MIC90 results are shown in Table 3.
Minimal Inhibitory Concentration (MIC) of CoPS isolated from a veterinary teaching hospital against antimicrobial agents.
| Species | S. aureus (n=18) | S. pseudintermedius (n=27) | ||||
|---|---|---|---|---|---|---|
| Antimicrobials | MIC (μg/ml) | |||||
| Range | MIC50 | MIC90 | Range | MIC50 | MIC90 | |
| Oxacillin | 0.25–>64 | 0.5 | 64 | 0.25–>64 | 0.5 | 64 |
| Cefoxitin | 2–32 | 4 | 32 | 0.5–16 | 0.5 | 4 |
| Enrofloxacin | 0.25–64 | 0.5 | 16 | 0.25–64 | 0.5 | 64 |
| Clindamycin | 0.25–32 | 0.5 | 32 | 0.25–32 | 2 | 32 |
| Vancomycin | 2–>64 | 2 | 4 | 2–4 | 2 | 2 |
Eleven strains were positive for the mecA gene (24.4%). Of these strains, six were obtained from the veterinarians, two from hospital surfaces, two from an owner and one from a canine. All methicillin-resistant strains harbored this gene.
Genotyping of strains by SmaI and PFGEThe PFGE profile (pulsotype) was obtained in 41 of the 45 strains. Four of the strains were not typable by SmaI enzyme digestion and PFGE, even when the electrophoresis was performed with thiourea. The untyped strains corresponded to one strain of S. aureus and three strains of S. pseudintermedius, all isolated from the canines. Among the pulsotypes identified there was high polyclonal diversity, displaying clonal relationship in two MRSA strains (MV1 and MV2) isolated from the veterinarians and in two S. pseudintermedius strains (C28 and C30) isolated from the canines (Fig. 1, supplementary material).
Statistical analysisThe percentage of isolated CoPS strains, and their susceptibility to methicillin from every source was analyzed. No statistical difference was observed in CoPS isolated from canines, veterinarians and owners; the only statistical difference was obtained with the percentage of methicillin-resistant strains isolated from veterinarians (6/8) and owners (1/8) (p<0.001).
DiscussionDifferent publications report variable percentages of nasal carriage of Staphylococcus aureus in humans, which vary from 18.5 to 27.7%22,33. In the present study a total of 16 CoPS strains were isolated from the 64 samples obtained from humans (25%). It should be noted that when separating the percentages obtained from owners and veterinarians, there was no statistical difference (p<0.35), in agreement with Kottler et al.24, who found no differences between the general population and people associated with health careers in terms of percentage of isolation of CoPS strains.
Two S. pseudintermedius human carriers were found in the present study. To our knowledge, this is the first national study documenting the carriage of this species in humans. Although this bacterium is adapted to living in animals, it is documented that S. pseudintermedius has some properties that make possible the presence of this microorganism in human skin; in fact, Latronico et al.25, describes a particular sequence type (ST-71) that adheres to the human corneocyte with the same ease as to the canine corneocyte. In general, the isolation percentages of this species in humans are low; however, the most modern and precise identification methods are likely to generate an increase in the number of cases of strains isolated from humans42, because, given its biochemical similarity, it can be misidentified as S. aureus7. This is particularly important in methicillin-resistant strains, where interpretation criteria and MICs defined for S. aureus can lead to MRSP isolates being defined as susceptible7. Nevertheless, transmission of mobile genetic elements such as SCCmec or other resistance determinants from S. pseudintermedius to other Staphylococcus species has been previously reported34.
Regarding canines, CoPS strains were isolated from 26 of the 40 patients sampled (65%), a percentage lower than that described by Muñoz et al.32, who reported 71.3%, despite analyzing only animals with skin conditions. In total, 26 strains were obtained, 10 from nasal cultures and 16 from the perianal area, in agreement with Hanselman et al.17, who reported that perianal colonization was significantly more common than nasal colonization. Of the strains isolated from canines, 22 (84.6%) corresponded to S. pseudintermedius and 4 (15.4%) to S. aureus, which is expected because the dog is the natural reservoir of S. pseudintermedius26.
With respect to S. aureus strains, the resistance to erythromycin is 38.9%, similar to that described by Rahman et al.36, while for S. pseudintermedius strains, resistance was detected in 37% of the strains (Fig. 2), similar to that described by Vigo et al.46, and Gröntal et al.16, who described 39.3 and 30.5% of resistance, respectively. The slightly higher percentage of resistance in S. aureus may be explained by the limited use of this drug in the veterinary clinical practice in Chile13. In fact, Awosile et al.4, described that erythromycin resistance is decreasing over time for CoPS isolated from dogs and Rumi et al.39, described a clear resistance increment to erythromycin in S. aureus.
With regard to clindamycin, 33.3% of the strains were resistant, which acquires great relevance when considering the publication by Hillier et al.18 that describes clindamycin as one of the first-tier antibiotics for the treatment of canine pyoderma, which is caused in the majority of cases by S. pseudintermedius26, demonstrating the importance of analyzing the behavior of these strains at a local level to establish the best treatment strategy. In this context, Vigo et al.46, stated that clindamycin should be empirically avoided and Rumi et al.39, described more than 40% resistance in Staphylococcus spp. Therefore, it is necessary to consider other treatment alternatives, such as topical chlorhexidine therapies, to avoid systemic antimicrobial therapy and decrease the emergence of methicillin-resistant strains that unnecessarily lengthen therapy6.
When considering resistance to fluoroquinolones by species, a higher percentage of resistance was found in S. aureus strains versus S. pseudintermedius strains (Fig. 2). Even though there is not a clear explanation of this, it agrees with the literature9, although a growing fluoroquinolone resistance rate was observed in Staphylococcus spp. recovered mainly from skin and ear samples in companion animals in Argentina39.
With regard to cephalosporins, 13.3% of the strains showed resistance to cefoxitin considering both species. It should be noted that none of the strains isolated from canines exhibited resistance (Fig. 1), which differs from Saputra et al.’s study40 that reported 12% resistance against cefoxitin. This difference can be explained by the number of strains analyzed in that study, which was highly superior to those in the present study (888 strains versus 45); therefore, it is likely that if the number of strains studied is increased the resistance percentages may change.
With respect to tetracycline, 15.6% of the CoPS strains were resistant. It is important to consider that no S. aureus strains exhibited resistance; however, 25.9% of S. pseudintermedius strains were resistant (Fig. 2). Moreover, Saputra et al.40, and Gröntal et al.14, reported higher percentages of resistance of S. pseudintermedius versus S. aureus, probably due to the more frequent use of this drug in veterinary settings5.
No resistance was found to doxycycline and amikacin and a lower percentage of resistant strains was found to gentamicin (2.2%), which coincides with that described by Gröntal et al.16, who reported low levels of resistance against doxycycline (6.1%), amikacin (0%) and gentamicin (6.6%). This low percentage of resistance against gentamicin was also reported by Ventrella et al.45, who explained it by the limited use of this aminoglycoside due to its high toxicity, which is also valid for doxycycline and amikacin13. Additionally, Rumi et al.39 reported that resistance to amikacin never exceeded 1% in Staphylococcus isolated from companion animals.
Regarding phenotypic susceptibility against oxacillin, nine methicillin-resistant strains were found (20%), all of them harboring the mecA gene. Two other strains, which showed phenotypic susceptibility to oxacillin, were also carriers of this gene (one strain of S. aureus isolated from an owner and a S. pseudintermedius from a canine), which could be explained by the lack of the promoter or other sequences that allow its expression49. This phenomenon of mecA-harboring strains and susceptible to oxacillin was already described in Chile in strains isolated from cats14. When considering their MICs, it should be noted that two strains from canines that were susceptible to oxacillin by the agar diffusion test were resistant by MIC; however, these strains did not carry the mecA gene and were susceptible to other β-lactams, thus were considered methicillin-susceptible and probably hyperproducers of beta-lactamases strains30.
Of the eight strains isolated from veterinarians, six were MRS (75%), which is statistically different from the 12.5% of methicillin-resistant strains isolated from owners (p<0.001). This agrees with Rodrigues et al.37; Pomba et al.34, and Kottler et al.24, who established that carrying methicillin-resistant strains could be considered an occupational risk. Regarding S. pseudintermedius, we found one veterinarian carrying a methicillin-resistant strain, which represents 4.1% of all the veterinarians sampled, similar to that reported by Febler et al.11, who described 3.6% of veterinary personnel carrying MRSP strains. No MRSP strains were isolated from owners, in accordance with Gómez-Sanz et al.15, who did not find any MRSP from 67 owners, and to the review published by Pomba et al.34, who described that colonization of MRSP in humans seems to be rare and transient. This is the first report in Chile of a human harboring MRSP.
Regarding the 10 hospital surfaces sampled, two MRSP were found, both from hospitalization cages, highlighting the importance of the environment in the possible transmission of these strains between humans and pets. In fact, Hogan et al.19, asserted that the search for MRS strains exclusively in humans is inadequate, since the environment is increasingly recognized as a reservoir of these strains.
The results obtained confirmed that the cefoxitin disk is not a good predictor of the presence of the gene in S. pseudointermedius strains, since the three strains of S. pseudointermedius that carry the mecA gene and are phenotypically resistant to oxacillin are susceptible to cefoxitin, which has been widely described in the literature7–9.
We did not isolate any MRS from canines, which is in line with the publication of Katakweba et al.22 This accounts for less than the 2.6% described by Kjellman et al.23 (2015) in Norway; 6.5% described by Menandro et al.31 in Italy; and 11.7% described by Couto et al.10 in Portugal, which could be explained by geographic differences, although it must be noted that these studies consider a much higher number of isolates than the 45 strains analyzed in the present study. Another important factor is that this study includes only healthy dogs, and the percentage of methicillin-resistance carriage is higher in patients that have chronic skin diseases34; therefore, it is relevant to consider diseased animals in future studies. In fact, in the aforementioned studies, Kjellman et al.23, who only considered healthy animals, described a lower percentage of MRS isolation compared to the studies conducted by Menandro et al.31, and Couto et al.10, which only considered animals with skin conditions.
Regarding the percentages of drug resistance profiles of the CoPS strains, these are much higher in methicillin-resistant strains versus methicillin-susceptible strains to non-β-lactamic antibiotics in both species analyzed, as reported by Gröntal et al.16, and Ventrella et al.45, highlighting 100% resistance against ceftriaxone, enrofloxacin, ciprofloxacin, clindamycin and erythromycin in the present study (Fig. 3). It should be noted that all the methicillin-resistant strains analyzed here were susceptible to amikacin, doxycycline and gentamicin.
When analyzing the vancomycin MICs, we found 2 resistant strains (both S. aureus methicillin-sensitive isolated from owners) and 4 other strains with intermediate susceptibility (2 MRSP isolated from surfaces and one S. aureus and one S. pseudintermedius isolated from canines, both methicillin sensitive). This finding needs to be analyzed in depth, but it will remain for further analysis, highlighting the great relevance that this phenomenon would have, since until now vancomycin resistance in Staphylococcus has not been described in our country, except for one publication that reported heteroresistance in humans44; therefore, it is still considered the first treatment alternative in cases of MRS in human medicine in Chile29.
In this study, the PFGE profile of 41 strains was obtained. As in our study, Kadlec et al.21 also reported that it was not possible to determine the PFGE profiles of various S. pseudintermedius strains. In the mentioned study, it was possible to determine the pulsotypes of the strains using the ApaI enzyme, which could not be done in the present study. The identified strains showed great pulsotype diversity, clonality was found in two strains of MRSA isolated from veterinarians and in two strains of S. pseudintermedius isolated from canines. This phenomenon of clonal diversity agrees with that published by Gagetti et al.12, in S. pseudintermedius strains isolated from ill canines and by Kjellman et al.23, in S. pseudintermedius strains from healthy canines. Due to the aforementioned clonal diversity, it is not possible to infer transmission of these strains between humans and pets; however, it is possible to find clones by increasing the number of strains analyzed, as described by Kotler et al.24, who analyzed 586 S. aureus strains and found 4 MRSA strains with indistinguishable strains in owners and their pets.
The main constraint of the present study is the limited number of strains analyzed. The samples were taken to work with 50 strains, and studies with more strains are needed to evaluate clindamycin resistance in a more representative manner. It was not possible to use the ApaI enzyme for a better characterization of all strains isolated. Furthermore, there have been more accurate methods for surface samples published after this study2.
ConclusionThis study suggests that veterinarians are at high risk of harboring methicillin-resistant CoPS and provides evidence that clindamycin may not be a good alternative for CoPS in the analyzed hospital, although more studies are needed in this area for better understanding the resistant dynamic in CoPS in Chile.
Conflict of interestThe authors declare that they have no conflicts of interest.
The authors appreciate the kind collaboration of all staff of Laboratorio de Investigación en Agentes Antibacterianos at Facultad de Ciencias Biológicas, Universidad de Concepción, and the Laboratorio de Microbiología Clínica Veterinaria of the Facultad de Ciencias Veterinarias y Pecuarias of the Universidad de Chile (FAVET). This study was financed by the Conicyt National Doctorate Grant No. 21141033-2014, and by the ANID Millennium Science Initiative/Millennium Initiative for Collaborative Research on Bacterial Resistance, MICROB-R, NCN17_081-2020.