Most cases of hepatocellular carcinoma (HCC) are able to be diagnosed through regular surveillance in an identifiable patient population with chronic hepatitis B or cirrhosis. Nevertheless, 50% of global cases might present incidentally owing to symptomatic advanced-stage HCC after worsening of liver dysfunction. A systematic search based on PUBMED was performed to identify relevant outcomes, covering newer surveillance modalities including secretory proteins, DNA methylation, miRNAs, and genome sequencing analysis which proposed molecular expression signatures as ideal tools in the early-stage HCC detection. In the face of low accuracy without harmonization on the analytical approaches and data interpretation for liquid biopsy, a more accurate incidence of HCC will be unveiled by using deep machine learning system and multiplex immunohistochemistry analysis. A combination of molecular-secretory biomarkers, high-definition imaging and bedside clinical indexes in a surveillance setting offers a comprehensive range of HCC potential indicators. In addition, the sequential use of numerous lines of systemic anti-HCC therapies will simultaneously benefit more patients in survival. This review provides an overview on the most recent developments in HCC theranostic platform.
As one of the most recurrent and lethal cancers with highest burden, hepatocellular carcinoma (HCC) ranked the fourth commonest cause of death worldwide, particularly in East Asia and Western Africa, due to an estimated 5-year survival rate of approximately 20% [1]. Latest Global Cancer Data calculated 906,000 new diagnosed cases accompanied with 830,000 deaths from live cancer in 2020 [2,3]. Chronic hepatitis B virus (HBV), hepatitis C virus (HCV), and alcoholic liver cirrhosis or non-alcoholic fatty liver diseases contributed to developing HCC [4]. The 5-year survival rate can exceed 70% in patients if early detection of HCC is achieved followed by adequate treatments. The World Health Organization (WHO) recommends the employment of Alpha-fetoprotein (AFP) as an early diagnostic standard for screening HBV-infected patients with cirrhosis, further verifying with the ultrasonogram, magnetic resonance imaging (MRI) or computed tomography (CT) scan. However, such diagnostic imaging for screening is not affordable and available in developing jurisdictions. Neither the European nor the American HCC surveillance guidelines have excluded the quantitative determination of serum AFP due to its low diagnostic accuracy [5,6]. Consequently, the needs for better serologic HCC examination with sufficient sensitivity and specificity are essential.
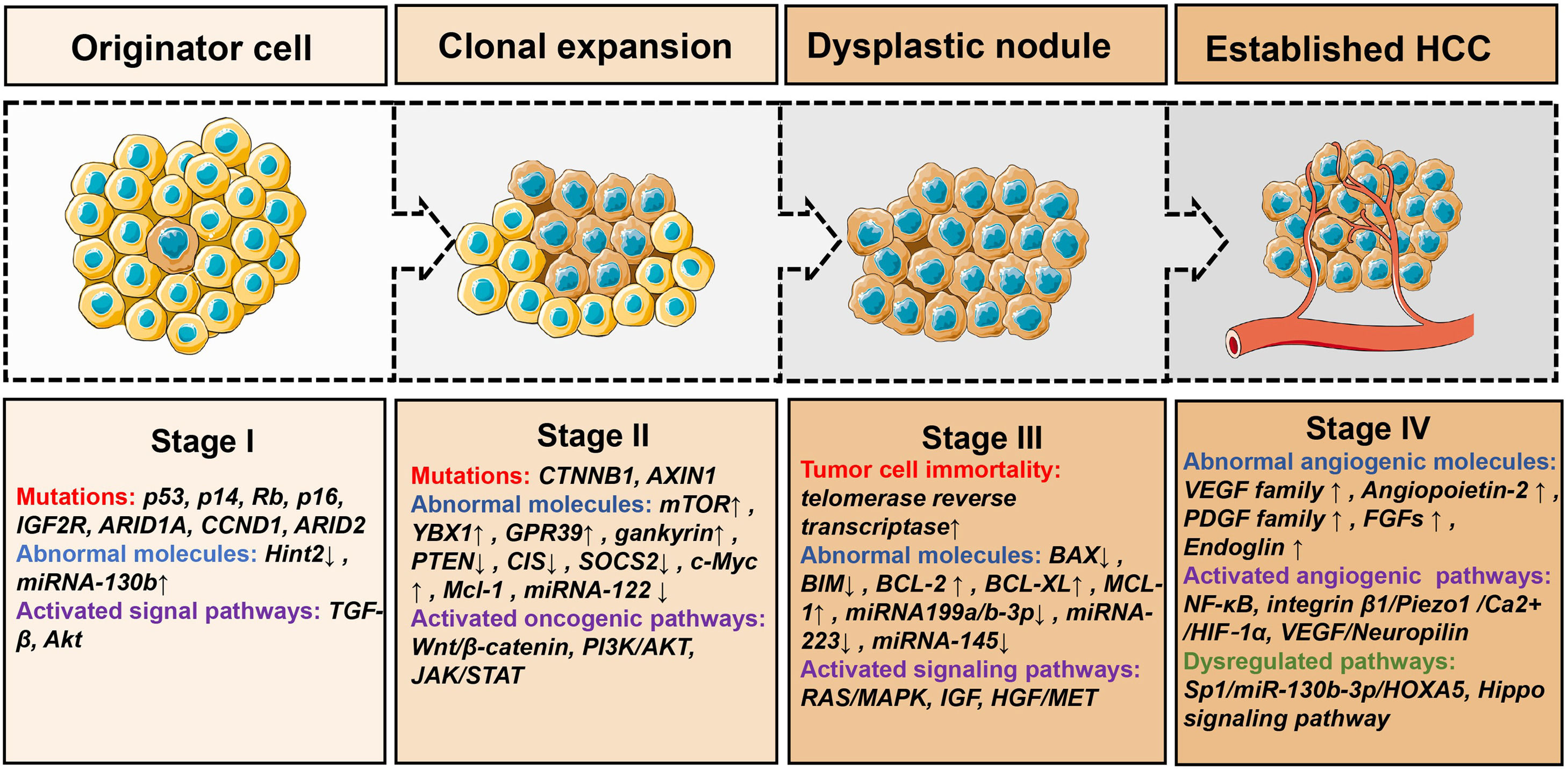
The pathogenesis of HCC still remains undetermined. The most common molecular changes in human HCC are shown in Fig. 1, which mainly include these four aspects: cell cycle checkpoint deregulation (p53, p14, Rb, p16 and IGF2R), resistance to apoptosis (Hint2, TGF-β pathway, insulin-receptor signaling and Akt pathway), activation of oncogenic signaling (Wnt/β-catenin and PI3K/Akt) and cancer cell immortality (the activation of telomerase) [7]. Multi-omic analyses of HCC tissue have revealed the somatic mutation accumulation, chromosomal abnormalities and epigenetic aberration in DNA methylation, for example, RB1, AXIN1, albumin, apolipoprotein B, CTNNB1, TP53, ARID1A and ARID2 [8]. The renin angiotensin system signaling pathways, for example, WNT/β-catenin, HGF/MET, PI3K/AKT/MTOR, VEGF, RAS/MAPK, IGF, PDGF and EGFR have been observed to activate through various growth factors, which lead to cell proliferation [9].
The pathogenesis and molecular changes of HCC. Stage I: This is the initial stage of hepatocellular carcinoma, and the main changes are somatic mutations [7-9,114]. Stage II: This is the stage of cancer cell cloning, and multiple mechanisms are involved [114–117]. Stage III: This is the progressive stage of hepatocellular carcinoma and cancer cells develop into dysplastic nodule [9,114,115]. Stage IV: This is the stage when the tumor has the ability to invade and metastasize [118–121]. ↑: up-regulate ↓: down-regulate.
A treatment plan for HCC is widely conducted to comply with the Barcelona Clinic Liver Cancer (BCLC) staging system, underlying progression and benefit assessment of medical interventions. In principle, resection, transplantation and local ablation are preferred in patients with early HCC, and transarterial chemoembolization (TACE) is preferred in patients with intermediate-stage HCC, and systemic therapy is recommended for patients with advanced unresectable HCC [10]. Early diagnosis and comprehensive treatment mainly by surgical excision are the key to the robust long-term therapeutic effect of HCC. However, therapeutic stratification of HCC patients remains challenging as half of the patients receive systemic therapy regardless of different disease stages [11]. Sorafenib presented the first-line systemic therapy approved by advanced inoperable HCC patients, and it took more than a decade before lenvatinib became the alternative first-line treatment recognized by US FDA. Second-line therapy options are composed of regorafenib, cabozantinib and ramucirumab [10]. Although these drugs have shown confirmed efficacies in clinical applications, therapeutic breakthroughs are still needed. With the deepening of studies, multiple biomarkers and molecularly targeted therapies have emerged in recent years. In addition, artificial intelligence technology brings more potentials for the diagnosis and therapy of HCC. Deep learning, in the era of big data, has been used to mine extensive biological information with diagnosis methods reported for a variety of cancer [12,13]. For instance, deep learning radiomics can describe the extent of axillary lymph node status in early-stage breast cancer and calculate the site and volume of primary tumors automatically in nasopharyngeal carcinoma [14,15]; deep learning of histopathological images offer genetic alterations in non-small cell lung cancer and Epstein–Barr virus infection in gastric cancer [16,17]; deep learning of retinal fundus images can stratify risk for chronic kidney disease and type 2 diabetes [18]. In this review, molecular-based HCC diagnosis and novel combination therapies on the current knowledge were summarized. In particular, we discussed the role of a deep learning assistant in the prediction of early recurrence of HCC.
2MethodsThrough literature reported on the prediction, diagnosis and therapeutics of HCC bedside reality, we carried out a systematic PUBMED search to compare and highlight relevant significant outcomes for future practical outlook. The publication date was covered from Jan of 2000 to May of 2023 and key concepts included “diagnostic biomarker”, “liquid biopsy marker”, “deep learning”, “systemic therapy”, “molecular targeted therapy”, “immune checkpoint inhibitor”, match with “hepatocellular carcinoma”.
All titles and abstracts were examined without article type restriction, and only publications regarding the following topics were selected: (1) practical biomarkers in liquid biopsy for HCC diagnosis, (2) deep learning in HCC diagnosis, (3) potential molecular targeted therapies aiming to improve the survival of patients with HCC or decreasing adverse events. Studies with ambiguous results in specificity and sensitivity of biomarkers for HCC diagnosis were excluded.
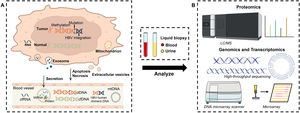
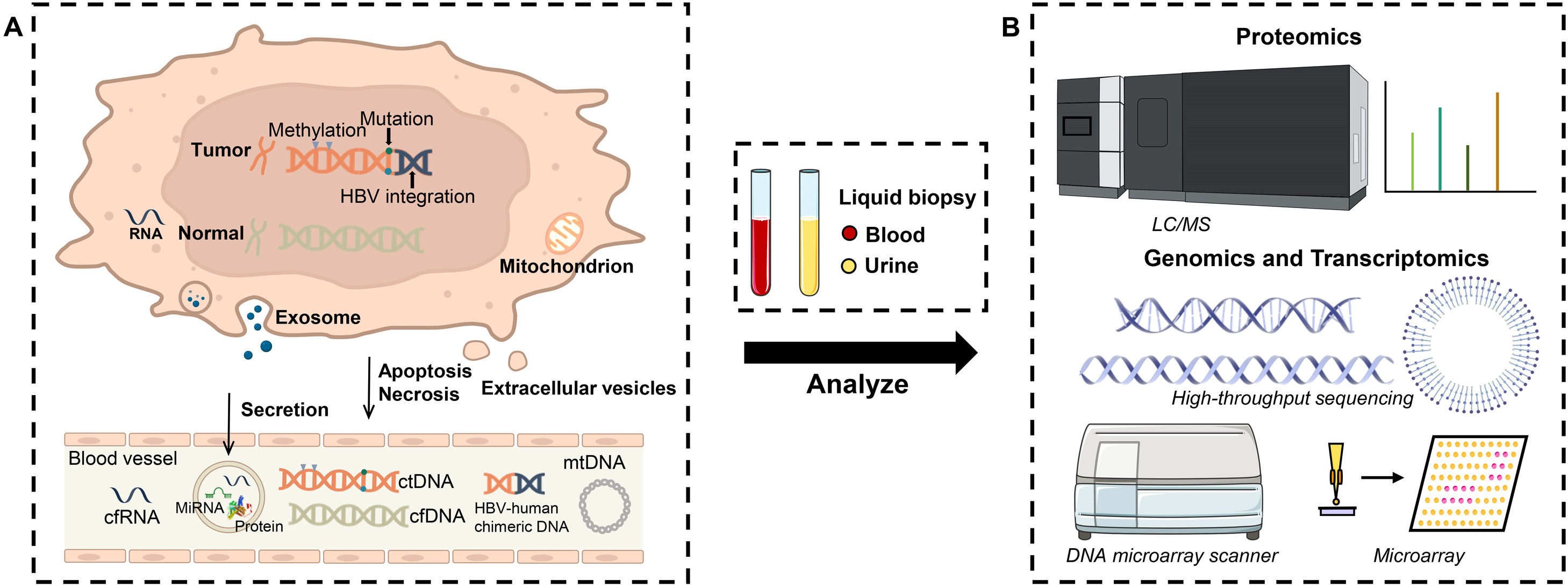
3Protein biomarkersIn HCC, liquid biopsy refers to the early detection and analysis of fragments of protein, DNA or microRNA (Fig. 2). The assessment of specific proteins released to determine the malignant lesions is a standard protocol for HCC diagnosis due to low cost, highly efficient, easy to attain and technical feasibility, for example, the familiar AFP and AFP-L3. However, their lower sensitivity and accuracy are the issue where major improvements are needed as there are numerous differences in diagnostic results among known protein markers.
Schematic illustration of working mechanism and device configuration for identifying variant or unknown HCC biomarkers. (A): Genetic and epigenetic changes were accompanied by HCC progress. HCC-associated proteins, DNAs and RNAs were continuously released into the bloodstream by secretion, apoptosis and necrosis, followed by capturing via liquid biopsy. (B): Subsequently, proteomic mass spectra, genomic and transcriptomic sequencings were utilized to discover new biomarkers in the collected samples for early diagnosis of HCC.
Hepatic derived AFP, as a fetal glycoprotein expressed in the embryonic liver, will be reactivated in liver disease or regeneration, showing a 73% sensitivity with a 92% specificity at early HCC detection [19]. Unfortunately, AFP predictive ability was highly dependent on the selected cut-off value during diagnosis, giving only a sensitivity of 61% at a cut-off value of 20 ng/mL and even dropped to 22% at a cut-off value of 200 ng/mL [20,21]. AFP began to fall out of favor for the HCC diagnosis in some countries, while it was still used as a golden standard to screen new biomarkers in early HCC surveillance. Recently, highly sensitive AFP-L3 fraction became the rising star in a large Japanese study, showing a sensitivity of 41.5% at a cut-off of 5% and a specificity of 85.1% in patients with AFP levels less than 20 ng/mL [22]. Furthermore, DCP referred to Des-gamma-carboxy prothrombin, derived by vitamin K Ab-sence II (PIVKA II), was measured as a predictor in Asia, providing a sensitivity of 48% to 62% and a specificity of 81% to 98% for HCC [23]. Compared with AFP, PIVKA II detection demonstrated some advantages in the diagnosis of HCC [24]. Another study found AFP combined with AFP-L3 and DCP maximumly improved HCC diagnostic accuracy [19]. To date, the Japan Society of Hepatology guidelines encouraged to evaluation of all mentioned biomarkers AFP, AFP-L3 and DCP as a package of surveillance programs and early HCC screen, however, such an approach has not yet been formally recognized by the major liver societies.
3.2Golgi protein-73(GP73), glypican-3 (GPC3), heat shock proteins (HSP) and epidermal growth factor-like domain-containing protein 7(Egfl7)Back in 2005, investigators considered a secretory Golgi phosphoprotein 73 (GP73) to be a biomarker, as its concentration positively correlated with HCC occurrence. GP73 significantly elevated its expression in HCC patients in comparison with those afflicted with cirrhosis. Following studies presented the AUC of GP73 (0.924) was higher than that of AFP (0.764) with a sensitivity of 88% and specificity of 87%, while AFP only showed 72% and 80% [25]. The combined detection method of GP73 and AFP had higher accuracy and sensitivity, resulting in a larger AUC than any individual use [26]. The combined use of GP73, Dickkopf-1 (DKK1) and AFP further showed the superiority with higher specificity and sensitivity [27].
GPC3, a heparin sulfate proteoglycan, has been proposed as a novel serum biomarker for HCC which is secreted by HCC-derived cell lines and upregulated in HCC tissues (elevated in serum of 40% to 53% of HCC patients) [28]. A meta-analysis of GPC3 showed a moderate accuracy to diagnose HCC with a mean pooled sensitivity of 82.5% and specificity of 57.8% [29]. Another potential biomarker for HCC is the heat shock proteins (HSPs) family, which is overexpressed in response to oncogenic stress in HCC. For example, the combination of HSP90 chaperones, AFP and thymidine kinase 1 (TK1) in HCC diagnosis increased the sensitivity of 89.24% and the specificity of 85.47% [30].
Egfl7 is an epidermal secretory protein that is specifically upregulated in HCC cells. Interestingly, the level of Egfl7 was only highly overexpressed in patients with early HCC, and hardly distinguished from the serum of healthy individuals. In spite of detecting the level of Egfl7 showing better sensitivity than AFP (77.4% vs. 65.3%), Egfl7 efficacy used as a serum detection marker for HCC remains to be further validated [31].
3.3Annexin A2, sAxl, and thioredoxin (TRXs)Serum Annexin A2 is also evidenced as a biomarker for HCC in 2009, due to the overexpression when binding calcium with phospholipid membrane in cirrhotic liver tissue and malignant hepatocytes [32]. Qiu et al. further confirmed that Annexin A2 mediated immune response in HCC tissues and highly promoted its levels compared with normal tissues [33]. In contrast with AFP, Annexin A2 performed comparable sensitivity (83.2% vs. 54.6%) and specificity (67.5% vs. 81.3%) at early detection of HCC, and reaching the maximum sensitivity and specificity at 87.4% and 68.3%, respectively, when using a combination of them [34].
Under HCC, Axl (a receptor tyrosine kinase) activated the PI3K/AKT pathway, Hippo/YAP signaling, TGF-β signaling and ERK/MAPK pathways, and further stimulated cell invasion and lymphatic metastasis, leading to proteolytically produce an 80-kDa soluble sAxl protein which secreted into the blood circulating [35]. In a study, the diagnostic ability of sAxl was verified, indicating that the concentration of sAxl obviously enhanced in the samples of HCC patient's serum with a sensitivity of 95% and specificity of 73.3% in comparison to those of AFP single diagnosis (sensitivity at 67.5%, specificity at 70%). In addition, the study found that the combined detection of sAxl and AFP achieved a very high AUC (0.914), with a sensitivity of 96.3% and a specificity of 72.5% [36].
Thioredoxins (TRXs) as thiol oxidoreductases could promote tumor cell proliferation and metastasis, correlated with tumor growth. Hence, circulating TRX significantly overexpressed in the liver homogenate, treated as another marker for HCC diagnosis [37]. To predict early-stage HCC with the largest tumor diameter of 2 cm or less, the sensitivity and specificity of TRXs were 74.9% and 87.5% with an AUC of 0.854, while AFP only gave 68.6% and 75.2% with an AUC of 0.720. Again, the combined prediction of TRX and AFP improved sensitivity to 81.3% and specificity to 93.4% with an increased AUC of 0.889, showing great clinical significance [38].
3.4Squamous cell carcinoma antigen (SCCA) and osteopontin (OPN)Squamous epithelia secreted sorts of subfraction of tumor-associated antigens, known as squamous cell carcinoma antigen (SCCA, a cytoplasmic glycoprotein), exhibiting comparative diagnostic potentiality in HCC. SCCA had a high sensitivity but low specificity (84% and 49%, respectively) that could be additional support to AFP [39]. Clinical studies pointed out SCCA-IgM immune complexes with a cut-off value of 95 AU/mL statistically differentiated in HCC patients (sensitivity, 92%; specificity, 98%; AUC, 0.932) [40]. Consequently, it would be used as either a single marker or a combination with AFP.
HCC-derived osteopontin (OPN) and its variants, known as glycol-phosphoproteins with a molecular weight of ∼44 kDa, stimulated several downstream signal pathways in HCC progression via integrin or CD44 receptors. In any stage of HCC, OPN performed the pooled sensitivity and specificity at 86% and 86%, respectively [41]. Referred to the case for early HCC prediction, OPN decreased its sensitivity to 75% but higher than AFP at 46% under the same conditions. Again, the combined utility of OPN and AFP achieved improved average accuracy (AUC, 0.81; sensitivity, 71%; specificity, 86%) [42].
3.5Midkine (MDK), dickkopf-1 (DKK-1), interleukin-6 (IL-6), vascular endothelial growth factor (VEGF), tumor-specific growth factor (TSGF) and transforming growth factor-beta 1(TGF-β1)With the advancement of Omics datasets, systems biology with high-throughput revolutionary techniques revealed more HCC associated protein markers with certain diagnostic significance, such as MDK, DKK1, IL-6, VEGF, TSGF and TGF- β1. MDK has higher sensitivity (88.5% vs. 74.4%), comparable specificity (80.6% vs. 84.4%) and better AUC (0.91 vs. 0.81) than single AFP [43]. DKK-1, a Wnt signaling regulator, is overexpressed in HCC tissues but relatively stable in nonmalignant hepatic diseases [44], giving a sensitivity of 93.6% and a specificity of 86.9% [43]. Using the cut-off-values of 13.5, IL-6 provided a sensitivity of 72% with a specificity of 73.5% for HCC. Combining IL-6, IGF2, AFP and platelet count further increased the sensitivity, which made up for the low sensitivity of AFP (AUC increased to 0.97, with 90% sensitivity and 85% specificity) [45]. High serum VEGF indirectly presented low efficacy and poor survival in HCC patients treated with TACE, regarding its prognostic value in HCC management [46]. Similarly, TSGF level also correlated with HCC staging advances, using a 62 U/mL cut-off assay with a sensitivity of 82% [47]. On the other hand, the enrichment signatures of TGF-β1 cytokine during hepatocarcinogenesis were increased, exhibiting a sensitivity of 68% and a specificity of 95% at 800 pg/mL cut-off [48]. However, these markers still lack efficient evidence to support their large-scale clinical utility.
4HCC-associated genesThe intricate interaction of genetic and nongenetic factors leads to the development of HCC. Genomic and epigenetic changes of circulating tumor DNA (ctDNA) and cell-free RNA (cfRNA) are potential biomarkers for HCC diagnosis [49]. DNA methylation is an epigenetic mechanism in HCC pathogenesis. MicroRNAs (miRNAs) which control cell proliferation, migration and invasion in the malignant tumor are often dysregulated in HCC. miRNAs associated with abnormal regulation are stable and detectable in liquid biopsy of HCC patients, and therefore they have high AUCs [50].
4.1DNA methylation - RASSF1A, HOXA9 and p16INK4ARASSF1A as a tumor suppressor gene control mitotic arrest, DNA repair and apoptosis, cell cycle and cell migration [51]. According to a meta-analysis, the detection of RASSF1A methylation has a sensitivity of 64.4%, a specificity of 87.5% and an AUC of 0.84, indicating that the potentiality of RASSF1A methylation as a biomarker with a higher level of moderate overall accuracy [52].
HOX genes play important parts in cell growth and apoptosis, cell differentiation and motility, signal conduction and angiogenesis. HOXA9 is a tumor suppressor gene that inhibits the growth and metastasis of tumor cells [53]. The recent methylation array study has shown significant hypermethylation of HOXA9 in HCC samples [54]. Compared to the detection utility in tissue samples, the detection utility of HOXA9 methylation for HCC in plasma samples has a better specificity (sensitivity, 73.3%; specificity, 97.1%; AUC, 0.835). And the combination of HOXA9 methylation and AFP (either parameter positive) has a more prominent sensitivity of 94.6% in plasma samples.
Studies have reported that hypermethylation of p16Ink4A leads to inactivation of p16Ink4A in HCC patients [55]. p16Ink4A as a tumor suppressor gene inhibits the cyclin D/CDK4/6 complex, thus induces cell cycle arrest [56]. A recent study assessed the detection of p16INK4A methylation by pyrosequencing, using circulating cell-free DNA from the serum specimens of patients with HCC, chronic liver diseases or healthy controls [57]. The detection of p16INK4A methylation has a sensitivity of 65.3%, a specificity of 87.2% and an AUC of 0.82.
4.2Nucleic acids - CXCR2, CCR2, and EP400As a subtype of the G-protein coupled receptor superfamily (GPCRs), CXC receptor 2 (CXCR2) is responsible for angiogenic activity and chemotaxis of neutrophils and endothelial cells. Using microarray analysis, Shi et al. identified three HCC-specific genes, CXCR2, CCR2, and EP400, showing individual diagnostic accuracy in HCC patients with 82.4%, 78.4% and 65%, respectively [58]. Moreover, Qiao et al. concluded that CXCR2 was an adverse indicator of overall survival (OS) and relapse-free survival (RFS) in cancer patients with the exception of digestive tract cancer and was associated with poorer prognostic [59]. CCR2 is involved in the pathological processes of a variety of liver diseases such as hepatitis, cirrhosis, liver tumors and others [60]. EP400 has been implicated in cell cycle control, apoptosis and development. In addition, EP400 tightly associated with the RFS, disease-free survival (DFS) and OS of HCC patients [61]. The combination of these three genes was a feasible diagnostic mean for early-stage HCC with a sensitivity of 72%, specific of 95% and AUC of 0.959. If including AFP in the profile, the accuracy improved to 0.985 (sensitivity, 86%; specificity, 95%) [58]. Nevertheless, high costs of either DNA methylation or microarrays limit their application for general population screening.
4.3MicroRNAsMicroRNAs are tiny, non-coding and highly conserved RNAs and play a considerable role in post-transcriptional regulation. Numerous studies have shown that miRNA expression profiles reflect the biological behavior of tumors. In recent years, researchers are dedicated to improving the diagnostic utility of miRNA-based biomarkers for HCC. At present, two miRNAs, miRNA-21 (sensitivity, 87%; specificity, 80%; AUC, 0.88) and miRNA-122 (sensitivity, 90%; specificity, 94%; AUC, 0.954), demonstrated especially high potential as diagnostic tools for HCC [62,63]. Moreover, the combination of AFP and miRNA-122 further improved the accuracy of HCC diagnosis (sensitivity 90%; specificity 100%; AUC, 0.980). Haibo Zhou's team developed a biosensor for the synchronous and high-sensitive detection of three HCC associated miRNAs, including miRNA-21, miRNA-122, and miRNA-223 [64]. Besides, they further validated this multiplexed biosensor in clinical detection, opening a new avenue for HCC diagnosis. In another study, exosome-derived miRNA-21, miRNA-96 and miRNA-122 were not only significantly superior to the plasma expressions of these three miRNAs in the aspects of specificity and sensitivity for HCC diagnosis but also showed considerable accuracy in distinguishing the HCC group (sensitivity, 96%, specificity, 98%; AUC, 0.996) [65].
Serum miRNA-15b and miRNA-130b are other potential miRNA-based biomarkers that are significantly upregulated in HCC patients [66]. miRNA-130b is expected to be a powerful prognostic predictor for clinical application as giving a sensitivity of 87.7%, a specificity of 81.4% and an AUC of 0.845 [67]. The sensitivity of miRNA-15b was high at 98.3%, but its specificity was unacceptable at 15.3% for HCC diagnosis. More miRNAs markers including miRNA-139, miRNA-182, miRNA-331–3p and miRNA-199a-3P were recently studied for their diagnostic utility in HCC patients of all stages, yielding good accuracy (AUC >0.7) with high sensitivity (>60%) and specificity (>80%) [68–70].
Numerous studies are currently investigating the feasibility of using a panel of miRNAs to achieve higher sensitivity and specificity in HCC diagnosis. For example, the combination of miRNA-25, miRNA-375 and let7f was able to distinguish HCC patients with a sensitivity of 97.9% and a specificity of 99.1% (AUC, 0.997) [71]. Besides, the combined use of miRNA-10a and miRNA-125b separated the HCC group with a sensitivity of 98.5%, a specificity of 98.5% and an AUC of 0.992 [71]. In another research, a panel of circulating miRNAs comprising miRNA-26a, miRNA-27a, miRNA-122, miRNA-192, miRNA-21, miRNA-223, and miRNA-801 showed appreciable potential for the differentiation of HBV-related HCC (sensitivity, 81.8%; specificity, 83.5%; AUC, 0.888) [72]. Recently, Lin et al. developed a serum miRNA-based model consisting of miRNA-133a, miRNA-143, miRNA-145, miRNA-29a, miRNA-29c, miRNA-192, and miRNA-505, achieving better diagnostic sensitivity (70.4–85.7%) than AFP (40.7–69.4%) at a cut-off of 20 ng/mL in the early-stage HCC [73]. All results indicated that the detection of a panel of miRNAs as a biomarker appeared to provide an accurate mean for HCC diagnosis.
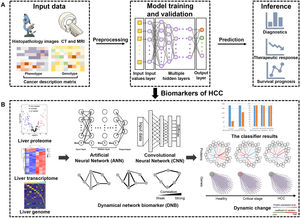
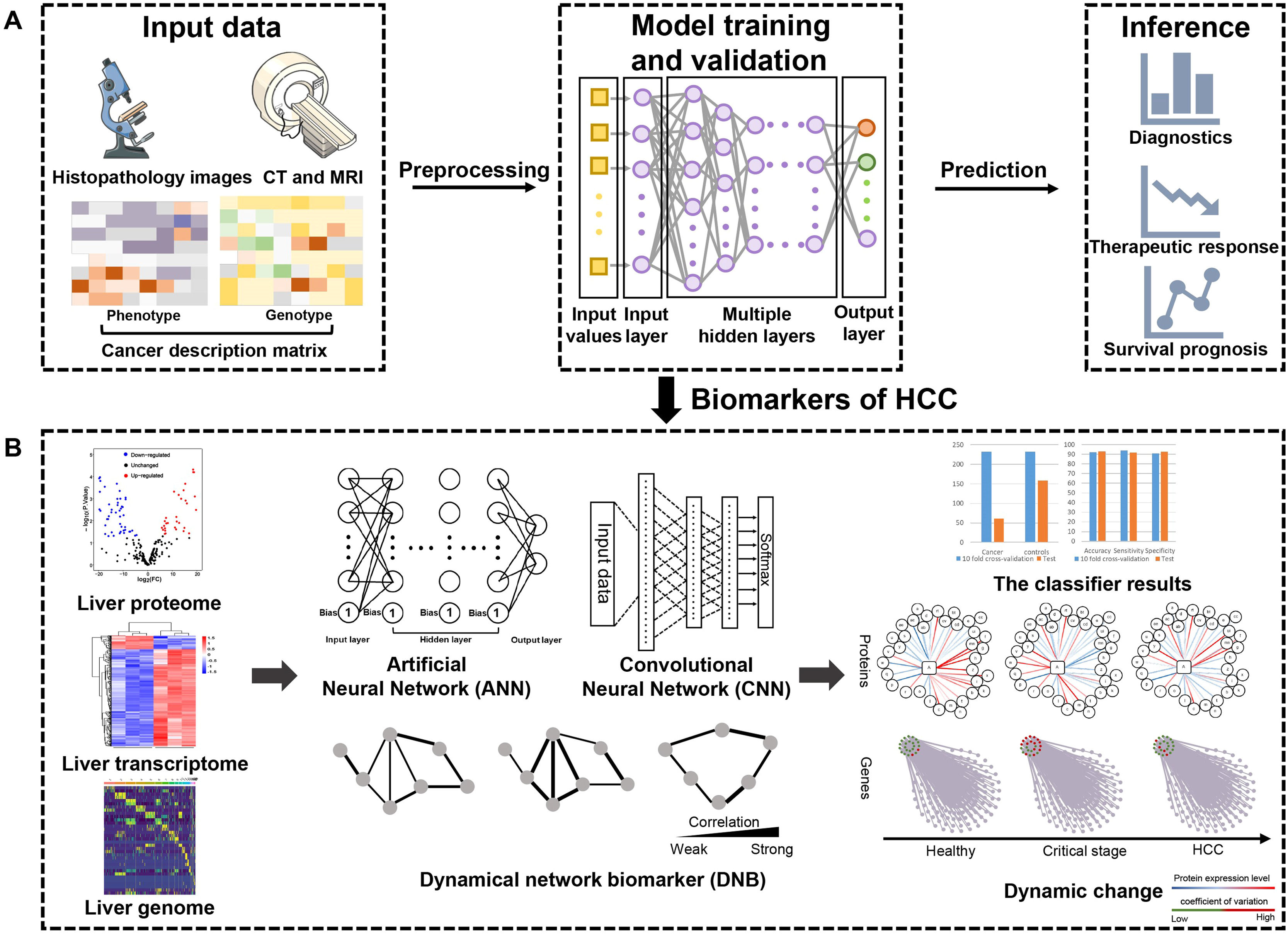
5Deep learningDeep learning, as a sophisticated AI technique, can process broad-spectrum medical data such as multi-omics datasets, radiologic datasets and histopathologic datasets [74]. It promotes the utilization of large data sets by directly learning the correlation between raw input data and target output. At present, numerous studies have reported the application of deep learning in cancer diagnosis, treatment and prognosis [12]. Patients with HCC generate a wealth of data, although significant heterogeneity exists. Deep learning has emerged as a unique opportunity to be applied to risk prediction, diagnosis, prediction and therapy in HCC [75], which is illustrated in Fig. 3.
Deep learning based multi-omics integration robustly identify HCC prognostic markers. (A): Input data including radiology images, histopathology section images, mRNA, DNA methylation and miRNA features are stacked up for autoencoder. Each transformed feature in the bottle neck layer of autoencoder is subject to single variate Cox-pH models, to choose the features associated with survival and identify survival-risk groups. (B): Those input features are ranked by ANOVA test F-values, those features that are in common with the predicting dataset are selected, then top features are used to build ANN and CNN models to predict risk labels for new samples. DNB model takes advantage of the dynamic nature of the data to predict critical transformations in complex diseases.
Numerous studies have explored the utility of deep learning based on imaging settings such as MRI, CT or histopathology. Liao et al. constructed a convolutional neural network (CNN)-based platform which used whole-slide images to distinguish tumors from adjacent normal tissues [76]. In addition to the use of an HCC imaging setting for pathological diagnosis, deep learning was also used to provide extensive biological information. For example, a recent study used an accurate deep learning score deriving from pre-therapeutic dynamic CT to assess survival in HCC patients who received TACE courses [77]. Imaging provides apparent information directly, but variations in the imaging of HCC are necessarily accompanied by numerous molecular transformations. Nowadays, growing deep learning models are focusing on molecular changes in HCC, which are not only used for the diagnosis of biomarkers but also provide therapeutic targets for drugs.
Over the past two decades, there has been an explosion in the acquisition of genomic and molecular data from bulk tissue and single cells. Recently, multi-omics related to HCC has had great application prospects for novel biomarkers [74,75]. And deep learning can be up to the computational challenge of high-dimensional data processing and analysis. Xie et al. built an ANN detection system based on nine genes expression of peripheral blood to distinguish patients with early-stage HCC and healthy individuals [78]. They combined peripheral blood, microarray analysis, the GeXP detection system and bioinformatics to build a diagnostic model, presenting a sensitivity of 98% and a specificity of 85% (AUC,0.943). Using ultra-deep sequencing and deep learning, Campo et al. found that the mean total entropy of HCC mitochondrial DNA (mtDNA) was 1.24-fold lower than that of non-cancer control mtDNA, suggesting that genetic diversity of intra-host mtDNA in blood could be a reasonable biomarker for the diagnostic detection of HCC [79]. Cheng et al. developed a nanobiosensing chip that acquired the HCC-relevant bioinformation in human serum through SERS detection [80]. Based on devised spectrum-based deep learning, this chip enabled label-free detection with simplified analysis procedures for point-of-care testing and low-cost (sensitivity: 90%; specificity: 92%). Besides, a dynamical network biomarker (DNB) model was invented to use the dynamic nature of biological data to predict the critical transformation of complex diseases or complex biological processes [81]. By using this model, the down-regulation of PLA2G6 and c-Myc was a warning sign of impending HCC. Moreover, a set of candidate genes was identified by using algorithms for dynamic network biomarkers to characterize different stages of HCC. Eventually, novel differential genes were valuable to distinguish cirrhosis which was the critical transition stage before oncogenesis [82]. Through the AI technique, Xu et al. discovered a lymphocyte migration-related kinase from HCC proteomics data, named as cytokinin 2 (DOCK2). DOCK 2 is highly expressed in HCC infiltrating T cells, which was crucial for effector T cell infiltration and anticancer function. Tumor cells with high expression of sulfonyl transferase 2B1 (SULT2B1) inhibited DOCK2 enzyme activity, acquiring resistance to immunotherapy such as immune checkpoint inhibitors, and thus accelerated the progression of liver cancer. This discovery took enlightenment to the clinical drug treatment of HCC [83].
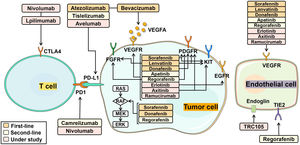
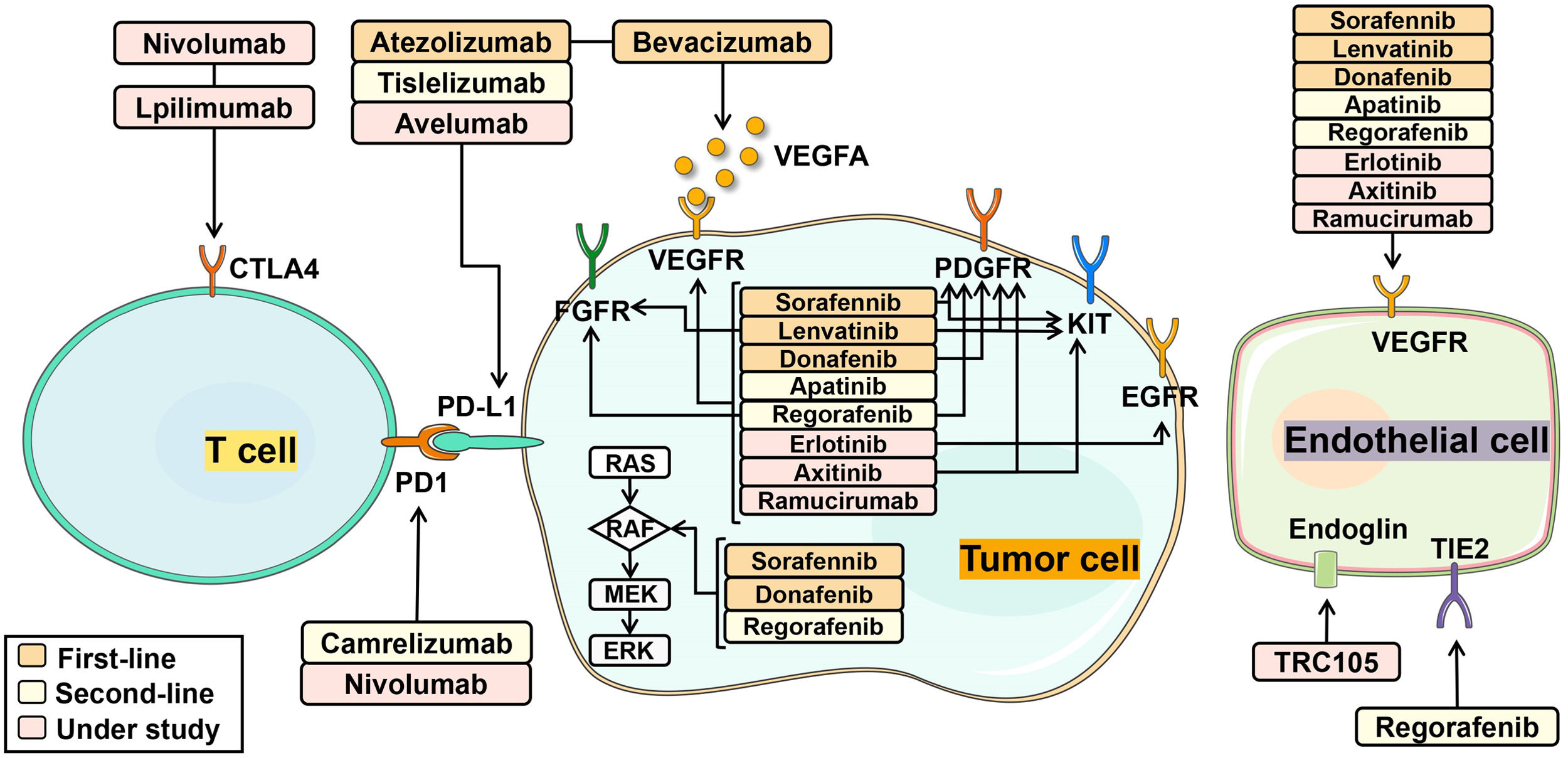
6Novel systemic therapiesHalf of HCC patients received systemic treatments regardless of disease stages. Systemic therapies included multi-kinase inhibitors, immune checkpoint inhibitors and monoclonal antibodies, by targeting specific molecules to delay cancer growth, progression and metastases [11]. Combining multiple drugs in HCC therapy produces a synergistic effect, enhancing the overall treatment response. Notably, single-cell sequencing analysis of HCC has proven the critical role of VEGF in driving tumor microenvironment reprogramming toward immunosuppression [84]. By targeting VEGF/VEGFR through different mechanisms, multi-kinase inhibitors and monoclonal antibodies effectively impeded tumor cell growth. Furthermore, the combination of multi-kinase inhibitors or monoclonal antibodies with immune checkpoint inhibitors also achieved improved efficacy by activating immune T cells [85]. HCC-targeted pharmaceutics with mechanisms of action is described in Fig. 4. In clinics, it is a new trend to combine molecular targeted therapy with cytotoxic chemotherapy drugs to jointly inhibit multi-targets in a complementary way.
The mechanism of molecular targeted therapies in HCC treatment. (1): First-line treatments: sorafenib, lenvatinib, donafenib, atezolizumab and bevacizumab. Sorafenib targets: VEGFR, PDGFR, KIT receptor, RAF kinase; Lenvatinib targets: VEGFR, PDGFR, FGFR, KIT receptor; Donafenib targets: VEGFR, PDGRF, RAF kinase; Atezolizumab targets: PD-L1; Bevacizumab targets: VEGFA. (2): Second-line treatments: apatinib, regorafenib, camrelizumab, tislelizumab. Apatinib targets: VEGFR; Regorafenib targets: VEGFR, PDGFR, FGFR, RAF kinase; Camrelizumab targets: PD-1; Tislelizumab targets: PD-1. (3): Drugs under study: erlotinib, axitinib, nivolumab, lpilimumab, avelumab, ramucirumab. Erlotinib targets: VEGFR, EGFR; Axitinib targets: VEGFR, PDGFR, KIT; Nivolumab targets: PD-1; Lpilimumab targets: CTLA-4; Avelumab targets: PD-L1; Ramucirumab targets: VEGFR.
Sorafenib, a tyrosine kinase inhibitor, plays a vital role in HCC therapy as a first-line treatment due to its multi-targeting functions. The phase III SHARP trial demonstrated patients treated with sorafenib owned a median overall survival (OS) of 10.7 months, 2.8 months longer as compared to the placebo controls (HR 0.69; P < 0.001) [86]. Sorafenib prevented tumor cell growth by inactivating the RAF-1, B-Raf and kinase activities in a Ras/Raf/MEK/ERK signaling pathway [87]. Although sorafenib showed its unique bioavailability than over dozens of other molecular drugs, the clinical efficacy of sorafenib had been largely limited by two problems: (1) The full mechanism of sorafenib resistance was still unclear; (2) New drugs combined therapy with sorafenib were essentially aimed to improve drug resistance and prolong survival. In a randomized phase II study of patients with advanced unresectable or metastatic HCC, investigators compared the effectiveness of sorafenib plus GEMOX (gemcitabine and oxaliplatin) combination therapy with sorafenib monotherapy. The results suggested that GEMOX combined with sorafenib was feasible for the first-line drug [88], with its median progression-free survival (PFS) improving to 6.2 months, an increase of 1.6 months over sorafenib monotherapy. However, the median OS was not apparently prolonged. An open-label and single-arm phase I study showed that TRC105 plus sorafenib had certain efficacy and safety (the median OS, 15.5 months; the median PFS, 3.8 months), treated in HCC patients who were sorafenib-naïve [89]. And the study was advancing to the phase II stage. Similarly, another phase I study found the three-drugs co-therapy including sorafenib, atorvastatin and metformin for advanced HCC patients was safe, exhibiting fewer sorafenib related adverse events (AEs) [90]. Conjoint therapy of sorafenib, metformin and atorvastatin could move forward in clinical trials.
6.2Lenvatinib monotherapy and its combinationsLenvatinib is a first-line treatment for liver cancer that has been approved by US FDA. Compared to sorafenib, lenvatinib inhibits a range of targets, including FGFRs, VEGFRs, PDGFRα and others. The phase III trial demonstrated that the median PFS of lenvatinib was twice that of sorafenib (7.4 months vs. 3.7 months) and the median OS of lenvatinib was as effective as sorafenib (13.6 months vs. 12.3 months) [91]. A phase Ib study of pembrolizumab plus lenvatinib for inoperable HCC patients showed that the objective response rates (ORRs) confirmed by independent imaging review (IIR) were 46.0% by mRECIST and 36.0% by RECIST v1.1 [92]. And the median durations of response (DORs) by IIR were 8.6 months per mRECIST and 12.6 months per RECIST v1.1. Additionally, patients treated with a combination therapy of lenvatinib and nivolumab achieved a better ORR (45.0% vs. 23.4%), PFS (7.5 vs. 4.8 months) and OS (22.9 vs. 10.3 months) compared to the lenvatinib monotherapy group [93]. However, this combination treatment remains to be proven by large prospective clinical trials.
6.3Atezolizumab plus bevacizumabAtezolizumab plus bevacizumab is a groundbreaking FDA-approved immunotherapy for first-line treatment in HCC patients. Atezolizumab specifically targets and inhibits PD-L1 and blocks the interactivity between B7–1 and PD-1, thus reactivating T-cell [94]. Bevacizumab, a monoclonal antibody, specifically binds to all VEGF-A subtypes to suppress angiogenesis and tumor proliferation [95]. Combination therapy of these two drugs has achieved remarkable success as a first-line therapy for HCC patients. According to a worldwide, open-label, phase III trial, this combination therapy not only improved OS at 12 months to 67.2% (67.2% vs. 54.6%), but also significantly prolonged median PFS compared to the sorafenib monotherapy (6.8 months vs. 4.3 months) [96].
6.4DonafenibDonafenib, a deuterium-labeled sorafenib, has been officially accepted by China's National Medical Products Administration in patients with HCC. Compared to sorafenib treatment, the donafenib treatment had a greater median OS (12.1 v 10.3 months) and fewer drug-related grade ≥ 3 AEs in patients (125 [38%] v 165 [50%]), and showed a similar tendency in median PFS between these two groups (3.7 months vs. 3.6 months) [97].
6.5Bevacizumab plus erlotinibErlotinib is also one of tyrosine kinase inhibitors through inhibiting EGFR signal transduction. A randomised, open-label, multi-institutional phase II study compared bevacizumab plus erlotinib therapy with sorafenib monotherapy to investigate the clinical efficacy and safety as first-line therapy for HCC patients [98]. Patients treated with bevacizumab plus erlotinib and sorafenib had essentially the same median OS (8.55 months vs. 8.55months; HR,0.92). The median event-free survival (EFS) was 4.37 months in the combination therapy and 2.76 months in sorafenib monotherapy. In terms of efficacy, no considerable difference was observed in the combination therapy and sorafenib monotherapy. Based on competing risks analysis, the sorafenib group had a higher incidence of interruptions for toxicity than the combination group (percentage of treatment completed for more than one cycle: bevacizumab plus erlotinib 87% vs. sorafenib 65%), suggesting that the combined therapy of bevacizumab and erlotinib was better than sorafenib monotherapy in terms of safety and tolerability.
6.6ApatinibApatinib is an antiangiogenic, small-molecule drug by selectively targets VEGFR2. A phase III study demonstrated that advanced-stage HCC patients who were refractory or intolerant to sorafenib or systemic chemotherapy, when treated with apatinib, had a noticeable prolonger OS (median 8.7 months vs. 6.8 months) and PFS (median 4.5 months vs. 1.9 months) compared to the placebo controls [99]. In addition, a nonrandomized, open-label, phase II study evaluated the utility of camrelizumab plus apatinib in HCC patients [100]. Both in first and second-line settings the combination of efficacy was shown to be robust for both ORRs (the first-line: 34.3%, the second-line: 22.5%) and median PFS (the first-line: 5.7 months, the second-line: 5.5 months) in advanced HCC.
6.7DovitinibDovitinib is an effective, multi-targeted tyrosine kinase inhibitor targeting FGFRs, VEGFRs and PDGFR, providing a dual mechanism of action anti-tumor activity including anti-proliferative and anti-angiogenic effects. In an open-label, randomized phase II trial among advanced HCC sufferers, the median OS and median time to progression (TTP) of dovitinib was similar to sorafenib (median OS: 8.0 months vs. 8.4 months; median TTP: 4.1 months vs. 4.1 months) [101]. A recent phase II study firstly used dovitinib as neoadjuvant therapy before local treatment of HCC. Twenty-four patients with early to mid-stage HCC received a 4-week course of oral dovitinib [102]. According to RECIST v1.1, 92% of patients had stable disease and 8% had partial response. Although some patients had a reduction in medication due to AEs, all had a significant reduction in intratumoural perfusion indices and patients who stopped treatment had a 29%−65% reduction in surviving tumors. With neoadjuvant therapy of dovitinib, all patients continued the planned local treatment without delayed toxicity.
6.8AxitinibAxitinib selectively targets VEGFR tyrosine kinase to inhibit receptor activities. In accordance with a multicentre phase II trial, the second-line treatment of axitinib is a safe therapy with moderate efficacy in advanced-stage HCC patients who have failed first-line sorafenib therapy [103]. According to RECIST v1.1 criteria, the axitinib group had a disease control rate of 62.2% and a response rate of 6.7%. In addition, a phase Ib trial proved the combination application of axitinib and avelumab (a PD-L1 inhibitor) as a first-line therapy for HCC patients had controlled safety and anti-tumor activity [104]. But their efficacy and safety were similar to other monotherapy or combination therapy. According to RECIST v1.1, this combination group had 5.5 months of median PFS and 14.1 months of median OS. 72.7% of patients received with axitinib plus avelumab had grade 3 treatment-related AEs (TRAEs), without showing grade 4 TRAEs and treatment-related deaths.
6.9Nivolumab plus ipilimumabNivolumab, the immunoglobulin (Ig) G4 monoclonal antibody against programmed cell death protein 1 (PD-1), revives the T cell fight against tumors via preventing the conjugation between PD-1 and its ligand PDL-1/2. Ipilimumab protects T-cell activation and proliferation via preventing the conjugation of CTLA-4 to ligands. The combination of these two drugs for advanced liver cancer patients who have failed sorafenib treatment has obtained accelerated confirmation from US FDA [105]. In the CheckMate 040 randomized clinical study, nivolumab combined with ipilimumab was administrated in HCC patients who had failed sorafenib treatment, with an observation of satisfactory results [106]. The ORRs were 32%, 27%, and 29% for the three experimental groups with different dose combinations of nivolumab and ipilimumab, respectively. Three groups shared similar response durations, median response times, and disease control rates as well as similar occurrences of treatment-related AEs. All patients responded early in treatment while a proportion achieved complete responses who had significantly lower tumor burdens. The group containing both nivolumab 1 mg/kg and ipilimumab 3 mg/kg had the longest median OS at 22.8 months and the longest median treatment duration of 5.1 months. Interestingly, in a virtual clinical trial using the Quantitative Systems Pharmacology (QSP) framework and a model calibrated with data from the clinical study CheckMate 040, similar consequences as in the clinical trial were obtained [107]. The overall response rates for patients treated with nivolumab, ipilimumab and the conjugation of nivolumab plus ipilimumab were 19.41%, 2.64% and 23.08%, respectively.
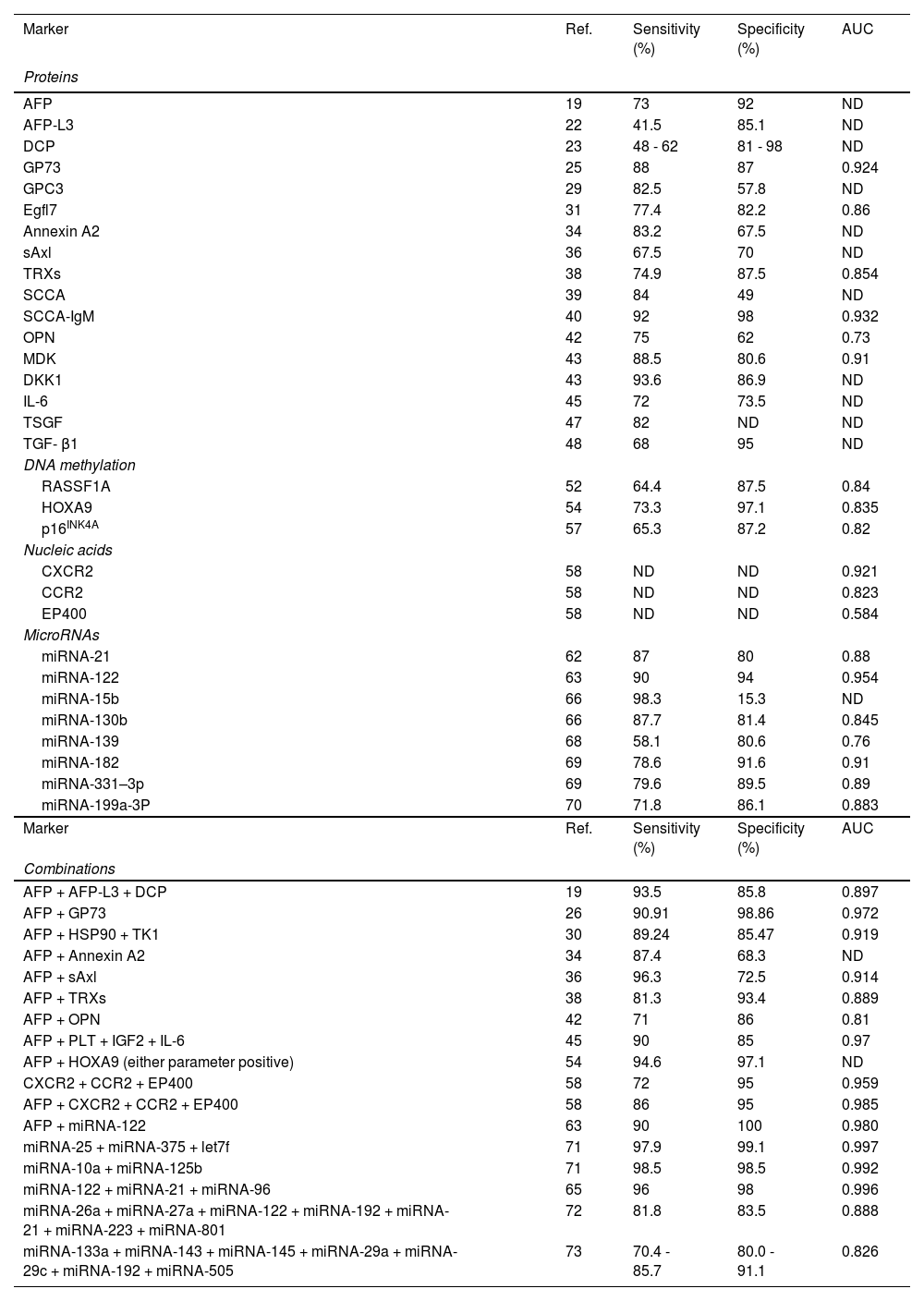
7DiscussionWe provided a brief insight into the developmental field of HCC theranostics. With more clarity, the diagnostic performance of all liquid biopsy markers mentioned above is given in Table 1. Not surprisingly, a single biomarker with both perfect sensitivity and specificity in all HCC cases is practically non-existent. With reasonable cost and easy availability, AFP still remains the gold-standard blood marker for HCC. Most diagnostic strategies depend on the combination of emerging biomarkers with affirmatory ones, particularly AFP, to improve diagnostic utility. Relevant studies showed that DCP, GP73, Annexin A2, sAxl, TRXs, OPN, CXCR2, CCR2, and EP400 achieved higher performance when combined with AFP. On the other hand, the newly identified class of HCC biomarkers (e.g., miRNA) combined with AFP may probably offer promising potential and benefit on improving diagnostic accuracy in the near future.
The Comparison of proteins, DNA mutations and microRNAs as biomarkers in Diagnostic Value of HCC.
AUC, area under the curve; ND, not determined; AFP, Alpha-fetoprotein; DCP, des-gamma-carboxyprothrombin; GP73, Golgi protein-73; GPC3, Glypican-3; Egfl7, Epidermal growth factor-like domain-containing protein 7; HSP, heat shock proteins; TRX, thioredoxin; SCCA, squamous cell carcinoma antigen; OPN, osteopontin; MDK, Midkine; DKK1, dickkopf-1; IL-6, interleukin-6; TSGF, tumor-specific growth factor; TGF- β, transforming growth factor-beta 1; CXCR2, CXC receptor 2.
Besides protein-based markers, genetic biomarkers appear to be the panel test that has appreciable clinical utility in HCC diagnosis. Indeed, the high cost of DNA methylation and array-based analyses may limit their application, therefore, a candidate-based approach will need to be standardized for effective implementation [108]. Moreover, the utility and reproducibility of miRNA data for HCC detection are limited by the result of miRNA levels via different extraction methods [109]. To improve the quantification of biomarkers, exosomes, discoid membranous vesicles with a diameter of 30–100 nm, have attracted attention in HCC studies. They are mainly from multi-vesicular bodies and can be released outside the HCC cells [110]. In a relevant animal trial, the combination of AFP, exosomes and circulating miRNA was more accurate in detecting HCC than AFP alone, suggesting this combination has appreciable potential as noninvasive novel biomarker [111,112]. Cell-free DNAs (cfDNA), based on their genomic and epigenetic changes, are promising as diagnostic biomarkers for HCC and monitoring of minimal residual disease due to the development of technologies such as droplet digital PCR and several next-generation sequencing strategies [49]. In addition, informative cfDNAs can provide accurate subtype classification.
The development of AI is promoting AI applications in all cancer fields and disrupting traditional research methods, owing to the high-dimensional dataset availability, deep learning architecture innovation and high-performance computing advances. For computer vision tasks such as image classification and face recognition, deep learning has achieved considerable success [113]. A great deal of research has been done in the early detection and diagnosis of cancer using deep learning. In the era of big data, the opening and sharing of databases such as NCBI, Ensembl and UCSC have gradually become one of the trends in the field of research. With more open clinical databases becoming available, discoveries will be made in combination with current analytical techniques.
The global burden of disease with HCC is increasing, and the improved implementation of primary and secondary prevention policies is essential to shorten morbidity and mortality. The five-year survival rate for hepatocellular carcinoma is approximately 20% [1], and most patients eventually develop into advanced-stage HCC so that only systemic therapies are available. Sorafenib and lenvatinib remain the most effective monotherapy [10], however, their median overall survival is still around one year. And a variety of tyrosine kinase inhibitors have been used as a first or second-line clinical treatment for advanced liver cancer, but their efficacy is limited. Currently, a combination of multi-kinase inhibitors, immune checkpoint inhibitors and monoclonal antibodies therapy that combine to inhibit two or more targets in a complementary manner to improve therapeutic efficacy is being studied. Some of them, such as the combination of atezolizumab plus bevacizumab, bevacizumab plus erlotinib and nivolumab plus ipilimumab, have yielded positive results and offered more potential treatment options for HCC patients. We suggest that the combination of nivolumab plus ipilimumab, which has shown a lasting effect in patients with advanced HCC and has been approved by US FDA, will be a rising ‘star’ in the HCC treatment landscape [105]. A phase III study is evaluating the efficacy of nivolumab plus ipilimumab in the first-line treatment of HCC patients. Combined with existing big data, artificial intelligence can be used to better discover therapeutic targets and the causes of drug resistance. These strategies will improve the standard of systemic HCC treatment by extending median OS beyond 2 years.
8ConclusionsAFP has been used as golden HCC biomarker for more than 60 years, and new series of biomarkers seem to be emerging. Various technologies such as genome-wide DNA microarray, RT-qPCR, proteomic and immunostaining studies have been applied to identify biomarkers for HCC diagnosis. There is a higher potential for combinations of these biomarkers to be of use in clinical practice, but their clinical application has to be precisely proved by large-scale validation studies in homogeneous ethnic and etiological populations. And the development of artificial intelligence also brings new prospects. From the examples mentioned, deep learning has shown appreciable capacity in cancer diagnostics. Improving data processing procedures and expanding training data by utilizing multiple sets of patients and samples largely increase the diversity of the AI. In terms of systemic therapy, combinations of multi-kinase inhibitors, immune checkpoint inhibitors and monoclonal antibodies therapy are emerging to compensate for the lack of monotherapy. It will broaden the clinical implementation of new therapeutic strategies against HCC.
Availability of data and materialThe data and material that support this review are openly available.
Author contributionsMiner Hu, Xiaojun Xia and Lichao Chen wrote the manuscript. Xudong Yao, Shudong Xia and Zhenhua Hu designed and revised the manuscript. All authors have approved this version of the article.




![The pathogenesis and molecular changes of HCC. Stage I: This is the initial stage of hepatocellular carcinoma, and the main changes are somatic mutations [7-9,114]. Stage II: This is the stage of cancer cell cloning, and multiple mechanisms are involved [114–117]. Stage III: This is the progressive stage of hepatocellular carcinoma and cancer cells develop into dysplastic nodule [9,114,115]. Stage IV: This is the stage when the tumor has the ability to invade and metastasize [118–121]. ↑: up-regulate ↓: down-regulate. The pathogenesis and molecular changes of HCC. Stage I: This is the initial stage of hepatocellular carcinoma, and the main changes are somatic mutations [7-9,114]. Stage II: This is the stage of cancer cell cloning, and multiple mechanisms are involved [114–117]. Stage III: This is the progressive stage of hepatocellular carcinoma and cancer cells develop into dysplastic nodule [9,114,115]. Stage IV: This is the stage when the tumor has the ability to invade and metastasize [118–121]. ↑: up-regulate ↓: down-regulate.](https://static.elsevier.es/multimedia/16652681/0000002800000006/v1_202310310350/S1665268123002417/v1_202310310350/en/main.assets/thumbnail/gr1.jpeg?xkr=ue/ImdikoIMrsJoerZ+w96p5LBcBpyJTqfwgorxm+Ow=)