In humans, microbial colonisation of the intestine begins just after birth. However, development of the normal flora is a gradual process, which is initially determined by factors such as genetic aspects, the maternal–foetal interaction, place and mode of delivery, early feedings strategies, and the use of antibiotics. Current knowledge on the significance and impact of the gut microflora on the development of the gut immune system indicates that a close relationship between allergic sensitisation and the development of the intestinal microflora may occur in infancy. Intestinal micro-organisms could downregulate the allergic inflammation by counterbalancing type 2 T-helper cell responses and by enhancing allergen exclusion through an immunological response.
As the prevalence rates of food allergy are increasing, hence there is an interest in understanding the reasons and in strategies to prevent or reduce the burden of the disease. The mechanisms leading to the incremented incidence of the allergic diseases are not fully understood. However, they are known to involve genetic factors as well as complex interactions between the host and the allergen exposure as well as other environmental stimuli on gut microbiota. Gut microbiota, which consists of microorganisms that live in the digestive tract, is the largest reservoir of human flora, having a continuous and dynamic effect on the host's gut and systemic immune system.1
Given the relative instability of the intestinal colonisation process during the first months of life, it is not surprising that any perturbation of this process may affect the microbiota, which may, in turn, have an impact on function and also potentially on the host's health. Delayed colonisation of the infant gut with commensal bacteria or alterations in the microbiota profile are suggested to be strong risk factors for the development of immune-mediated chronic disorders such as allergic diseases. Original research studies published in English between 1985 and 2012 were selected (Pubmed and Scopus). Computer searches used combinations of key words relating to “food allergy”, “microbiota” and “infancy”. In addition, the reference lists of the retrieved articles helped in the search for other relevant articles which were not found during the initial search procedure. Thus, 37 studies were selected and discussed here (15 randomised controlled trials, 10 cross-sectional, 4 case control, and 8 population based studies). The potential factors which may bias the findings of this review are the restriction to articles in English, together with database, and citation bias.
The aims of this review are to define potential factors which modify the intestinal microbiota of infants, to characterise the key mechanisms and their role in the development of food allergy.
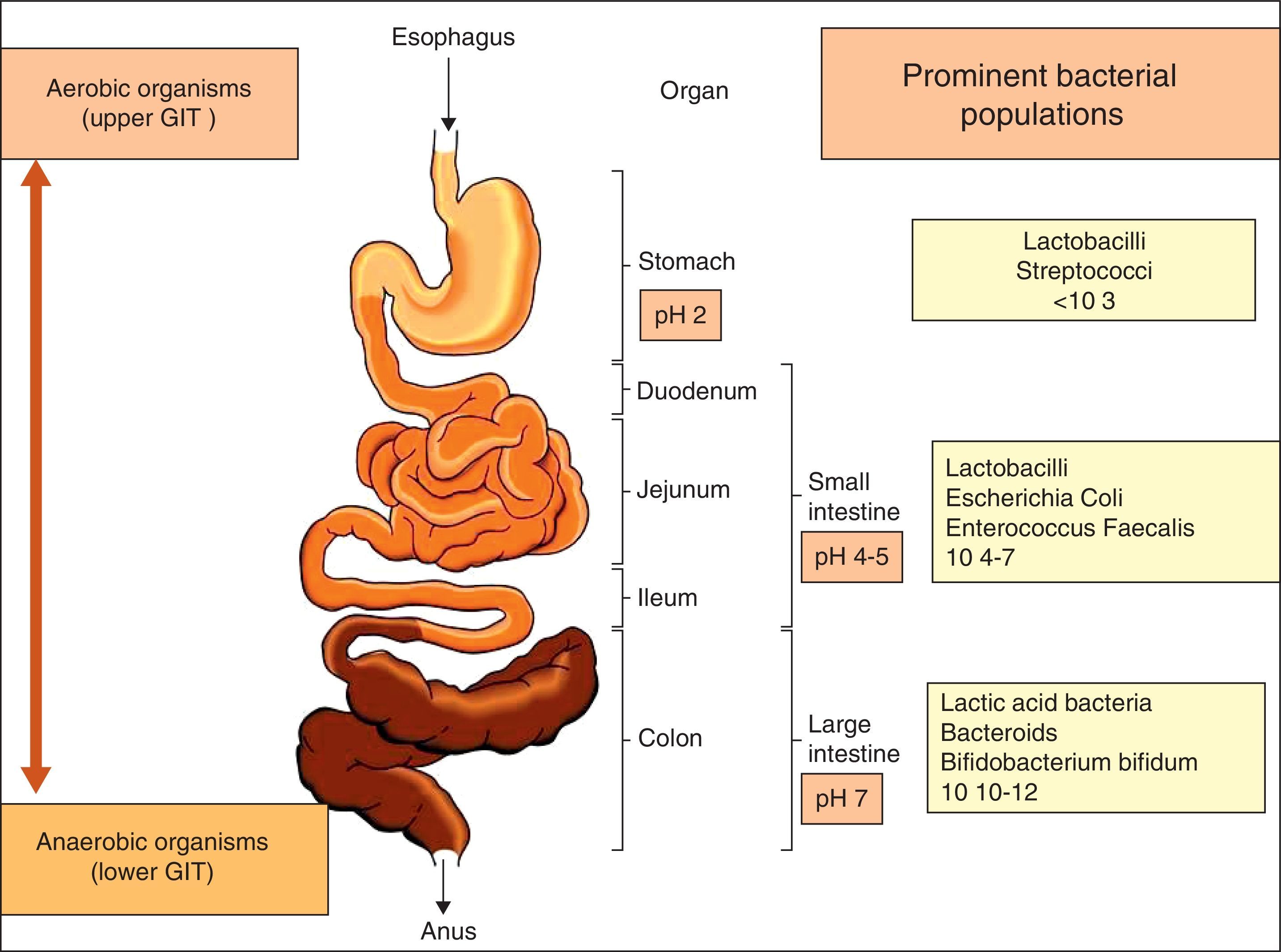
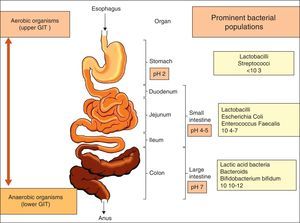
The gastrointestinal microbiota: diversity and propertiesNinety per cent of the human body is composed of prokaryote cells, and this group of microorganisms is known as microbiota.2 Microbiota is concentrated in the gut and plays a key role in the preservation of the host's health. The relationship between gut flora and humans is not merely commensal (a non-harmful coexistence), but rather a mutualistic relationship.3 The gastrointestinal tract, the largest immunological organ in the body, is in constant interaction with a high load of enteric flora and food antigens, resulting in a sustained state of low level physiological intestinal inflammation, protecting the host against severe infections but which allows immunological tolerance to endogenous and foreign antigens.4 The use of modern methods in molecular biology has provided further insight into the diversity of intestinal microbiota, which comprises hundreds of different species 5 that form a complex and highly interactive biomass (microbiome) of at least 1014 bacteria within the human gastrointestinal (GI) tract (Fig. 1). This microbiome contains more than 100-fold more genes than the human genome. The composition of the microbiota differs not only along with the length of the GI tract, but also cross-sectionally with different populations inhabiting the GI mucosa and lumen.6 The most common anaerobic genera in terms of concentration within the GI tract are Bacteroides, Bifidobacterium, Eubacterium, Fusobacterium, Clostridium and Lactobacillus. Among the facultative anaerobes are the Gram-negative enteric bacteria (Escherichia coli and Salmonella spp.), the Gram-positive cocci (Enterococcus, Staphylococcus and Streptococcus) and fungal species (predominantly Candida albicans).7
Another parameter which seems to contribute to the gut diversity is atopy. Dysfunction at the gastrointestinal barrier might contribute to an aberrant or exaggerated inflammatory response. Animal studies have shown that increased intestinal permeability is associated with food allergy.8,9 In addition, studies have also shown that early colonisation with potentially more pathogenic bacteria such as Clostridium difficile and Staphylococcus aureus is more likely to occur in children who go on to develop allergy. In contrast, lactic acid bacteria (Lab) and Bifidobacteria (Bfdba) are found more commonly in the composition of the intestinal flora of non-allergic children (Table 1). The enhanced presence of these probiotic bacteria in the intestinal microbiota seems to correlate with protection against atopy, by contributing to the production of T helper-1 immune responses, which may in turn block or prevent T helper-2 allergic responses in atopic disease.10 This approach may help to create optimal conditions for orienting T helper-2 polarised neonatal immune responses towards a positive T helper-1/T helper-2 balance. Furthermore, a slow functional maturation of the gut microflora, as measured by faecal levels of Short chain fatty acids (SCFAs: intermediates and end products of microbial action on endo- and exogenous, dietary as well as host derived components, mainly by anaerobic bacteria in the intestine) is associated with allergy, contributing to the development of microbial diversity.11
The diversity of gut colonisation according to feeding and atopy predisposition.
| Breastfed infants | 1st week of life: Bifidobacterium (infantis, longum, breve)33 | After weaning: Resembling the microbiota of adults with increased Bacteroids, Veillonela and Fusobacterium (yoshioka) |
| Lactobacillus (acidophilus)33 | ||
| Formula fed infants | Bifidobacterium (lower levels)9 | |
| Coliforms9 | ||
| Bacteroids9 | ||
| Atopic children | Clostridia (higher levels)8 | |
| Bifidobacterium (lower levels)8 | ||
| Non-atopic children | Lactobacillus (higher levels)9 | |
| Bifidobacterium (higher levels)8 |
As the gut microbiota is thought to drive the postnatal development of the immune system,12 the first months of life seem to be of major importance, as it is a ‘critical window’, during which the infant gut microbiota might influence key immunological events that alter allergic sensitisation.13 At this stage, the maturation of the immune system is not yet completed and is still building up immune tolerance against food and microbial antigens.14 Namely, the first oral exposure to food antigens results in the induction of T cells, which may mediate unresponsiveness to the systemically administered food antigen and hence lead to oral tolerance.15 During the crucial age of early infancy, an aberrant array or insufficient number of intestinal micro-organisms may not be able to potentiate the immature gut barrier or to counterbalance a Th2- skewed cytokine profile, i.e. to reduce two major risk factors for allergic sensitisation efficiently enough to avoid food allergy.
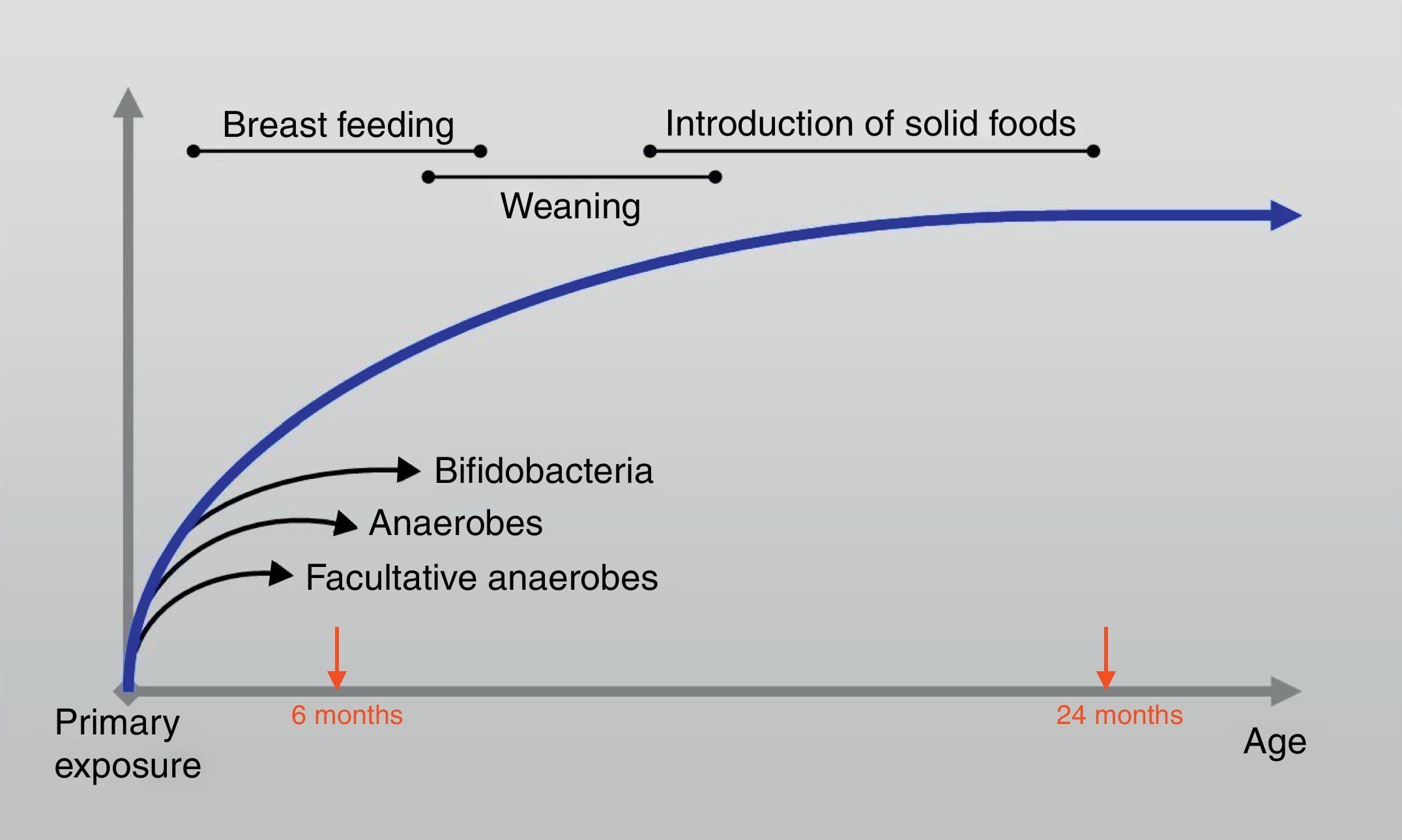
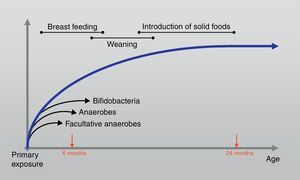
At the age of one month colonisation is complete and although the composition may fluctuate, large shifts in the composition do not occur until weaning.16 Furthermore; the intestinal microbiota evolves during the first two years of life towards a stable and definite future adult pattern that presents in adulthood (Fig. 2). During bacterial colonisation of the ileum and colon after birth, appropriate microbiological stimulation is essential to correct the balance of a skewed T helper-2 immune response predominant in neonates. In detail, the Th2 phenotype leads, however, to the stimulated production of IgE by B cells and thus increases the risk for allergic reactions through activation of mast cells. Microbial stimulation early in life will reverse the Th2 bias and stimulate the development of a Th1 phenotype and stimulate the activity of Th3 cells.17 Their combined action will lead to the production of IgA by B cells. IgA contributes to allergen exclusion and will thereby reduce exposure of the immune system to antigens. Cytokines produced by the Th1 phenotype will also reduce inflammation and stimulate tolerance towards common food antigens. 18
Similarly, infants with allergic disease have reduced numbers of beneficial bifidobacteria species and increased numbers of pathogenic Clostridia and staphylococci compared to non-allergic infants.19 Moreover, these changes occur prior to the onset of allergy, suggesting a causal relationship between microbiota and healthy immune responses and strengthening the importance of the intervention in the modulation of the gut microbiota during the infancy. Specifically, there are data to suggest that an aberrant microbial composition in the gut such as inadequate bifidobacterial biota may deprive the developing immune system from counter regulatory signals against T helper 2 mediated allergic responses.18,20
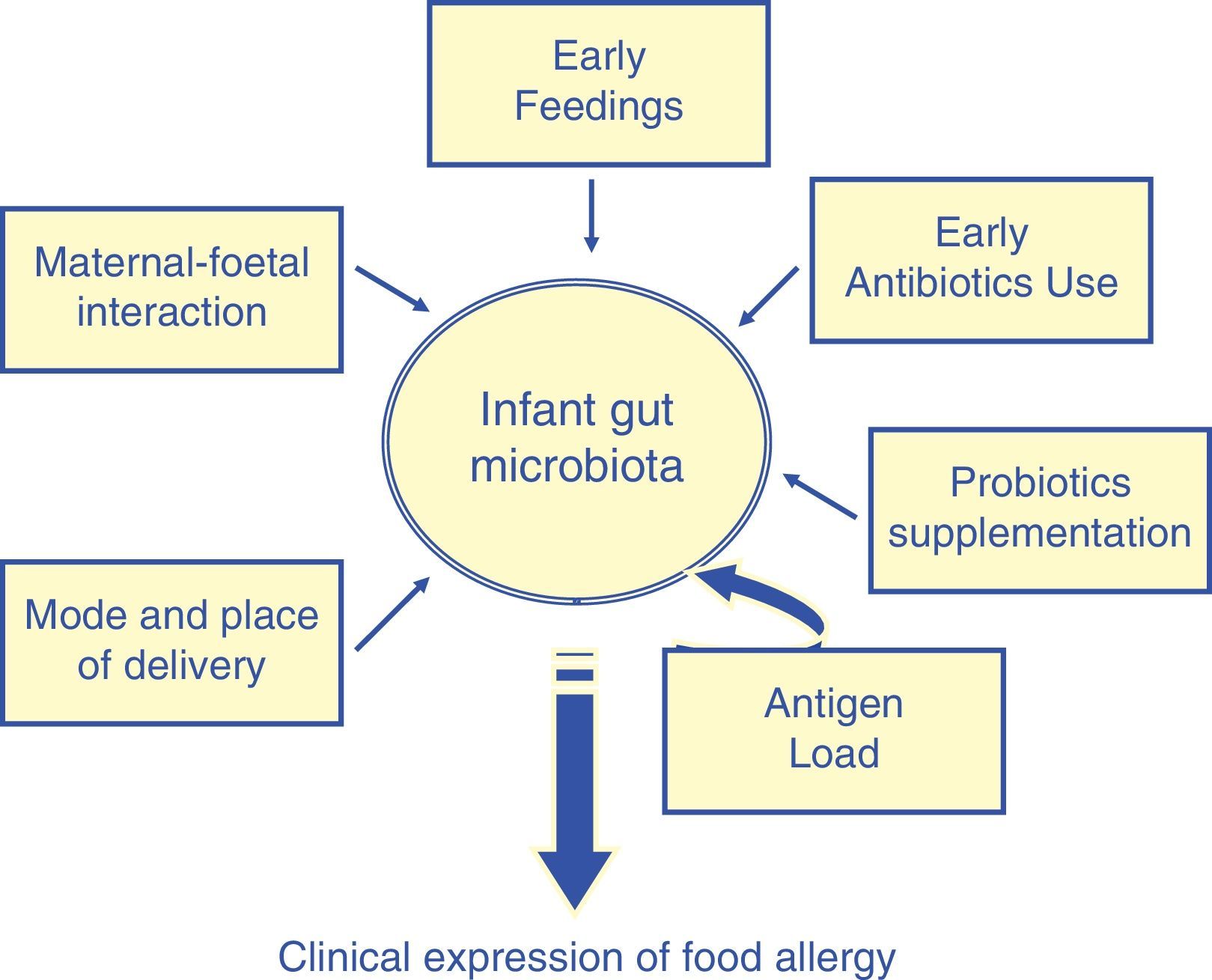
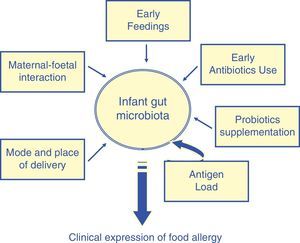
A wide spectrum of factors, such as the maternal–foetal interaction, place and mode of delivery, early feedings strategies, use of antibiotics,21 may promote the formation of ‘abnormal’ microbiota in infancy (Fig. 3).
Maternal–foetal interactionAt birth, the neonate leaves a germ-free intrauterine environment and enters a highly contaminated extrauterine world. The maternal flora constitutes the predominant source of initial colonisation. A part of it is transferred to the infant, influencing further intestinal microbiota development. Within the first few hours of birth, the process of intestinal colonisation takes place. The first bacteria colonising the neonatal colon are thus E. coli and various Enterococcus species 22 (Fig. 2). Obligate anaerobes follow. There is mounting evidence that the gut microbiota acquired during the early postnatal period is required for the development of regulatory T cell (Treg) population, which prevents the sensitisation to allergens. 20,21
It has been demonstrated recently that allergic mothers may transfer B. adolescentis species to their infants more often than healthy mothers,23 leading to aberrant compositional development of the microbiota, which may predispose infants towards allergic diseases.19 Recent discoveries suggest two new sources of intestinal microbes: bacteria in breast milk and microbes potentially present in the amniotic fluid before birth.24 Specifically, colostrum and breast milk are continuous sources of commensal, mutualistic and potentially probiotic bacteria to the infant gut.25 Breast milk bacteria may also participate in the correct maturation of the infant immune system since some strains are able to modulate both natural and acquired immune responses in mice and humans.26
Mode and place of deliveryCaesarean section consists in a perinatal risk factor for an increased risk of atopic disease, as shown in 2 meta-analyses.27,28 including food allergy. In a recent study, we showed that birth by caesarean section is associated with asthma and atopic sensitisation in children aged 8–9 years.29 The association of caesarean delivery with atopic sensitisation seems to be more pronounced in children with a family history of allergy. It has been suggested that the neonatal GM composition has an intermediate role in the relationship between delivery characteristics and the development of atopic disease. This is based on the fact that children born by means of caesarean section are not exposed to their mother's vaginal and faecal bacteria, unlike children born vaginally. In a recent study on the bacterial composition of the skin, oral mucosa, and gut, vaginally delivered infants harboured bacterial communities that were most similar in composition to the vaginal communities of the mothers, whereas infants delivered by means of caesarean section lacked bacteria from their mother's vaginas.30 Initially, children born by means of caesarean section are colonised by bacteria originating from the hospital environment and non-maternal skin bacteria,30,31 resulting in a delayed colonisation of the gut by beneficial bacteria and a different bacterial composition compared with that seen in vaginally delivered infants. Infants born by means of caesarean section have less frequent colonisation with lactobacilli and an especially lower abundance of Bifidobacterium and Bacteroides species. In contrast, they are more frequently colonised by C. difficile.15,32 These gut microbiota alterations are apparent within the first week of life preceding clinical symptoms, thus suggesting their causative role in allergic disorders.12,33 Furthermore, caesarean section is associated with increased IL-13 and INF-γ in neonates, which may contribute to the development of atopy.34
The place of delivery has an additionally important effect on the establishment of the GM composition because infants born vaginally in a hospital have a different GM composition, including more C. difficile, compared with home-born infants.15,30 Moreover, in a recent prospective birth cohort study, colonisation with C. difficile was associated with an increased risk of asthma at 6–7 years of age and wheeze, eczema, and sensitisation to food allergens until the age of 6–7 years.34 Furthermore, in children with a positive family history of atopy, vaginal home delivery was associated with a decreased risk of asthma and sensitisation to food allergens when compared with vaginal hospital delivery. Hence, it was for the first time demonstrated that not only the mode but also especially the place of delivery play an important role in the development of atopic manifestations in childhood.
The early feedings: what makes the difference?Regarding the age of the host in humans, it has been proposed that an immature intestinal barrier with increased permeability facilitates priming the humoral and the cell-mediated immune responses.35 The initiation of nutrition intake via the oral route shapes the early gut microbiota, and the healthy breast-fed infant can be considered here as a model of infant feeding. Breast feeding continues to enhance the original inoculum by the introduction of specific Lab, Bifidobacteria (Bfdba) and bacteria from the mother's skin, all of which enable the infant gut microbiota that is dominated by Bfdba. Breast milk also contains plentiful indigestible oligosaccharides, which pass through the whole intestine and promote the growth and activity of commensal bacteria, composed mainly of Bfdba.36 Molecular methods indicate that lactic acid-producing bacteria may account for <1% of the total microbiota, whereas bifidobacteria can range from 60% to 90% of the total faecal microbiota in breast-fed infants. These bacteria set the basis for gut microbiota development and modulation. The greatest differences between breast-fed and formula-fed infants appear to be in Lab and Bfdba colonisation. Usually, Bfdba appear after birth and, within a week, are reported as the dominant bacterial group, with Bifidobacterium (Bfdbm) infantis/longum/breve being the most common species in breast-fed infants.37 In addition, Lactobacillus (Lctbs) acidophilus is the most common Lctb in the faeces of breast-fed infants (Table 1). Formula-fed infants, on the other hand, tend to have a flora that is more complex, consisting mainly of Coliforms and Bacteroides, with a significantly lower prevalence of Bfdba.9 It was recently 38 demonstrated that the interindividual bacterial composition of the colonic flora in hospitalised preterm infants was markedly reduced compared with a large interindividual diversity in breast-fed full-term babies. Hence, the differences constantly reported between the gut microbiota of breast-fed and formula-fed infants may also lie in differences in the microbiological composition of human milk versus infant formula 9,12 above the bifidogenic oligosaccharides, in particular its 2-linked fucosylated oligosaccharides, in breast milk. After weaning, the introduction of solid foods and withdrawal of breast milk coincide with the next major microbiota changes in the colonisation process, although the impact of this phase is largely unknown. 39 Later in life, Bifidobacterium adolescentis becomes more common.12 Then, the microflora of children begins to resemble that of adults, with increased Bacteroides, Veillonella and Fusobacterium39 (Table 1).
AntibioticsThe major effects of antibiotic treatment on the microbiota are the most direct at killing a large proportion of the microbiota and the indirect effect of decreasing colonisation resistance within the GI tract. Colonisation resistance is a multi-faceted mechanism whereby obligate anaerobic microbiota inhibit the overgrowth of potentially harmful exogenous or endogenous microbes.40 Interestingly, changes in the microbiota populations can persist months after cessation of antibiotic therapy and can result in long-term decreases in beneficial anaerobic organisms (Bifidobacterium, Lactobacillus, Bacteroides) and increases in potentially harmful microbes (Gram-negative aerobic enteric bacteria, the anaerobe C. difficile and the yeast C. albicans).41,42 A number of studies have identified a correlation between early antibiotic use in children and the subsequent development of allergy/asthma.43,44 These results are consistent with the evidence derived from national trends of antibiotic use vs. incidence of allergic disease in industrialised (high atopy, high antibiotic use) vs. developing countries (low atopy, low antibiotic use).45,46 Moreover, studies have also concluded that rates of allergy among anthroposophic children (including a diet high in probiotics, fermented foods and restrictive use of vaccines, anti-pyretics and antibiotics) are also significantly lower.47 However, a study investigating anthroposophic children revealed that the use of antibiotics early in life was significantly associated with the development of asthma,48 which indicates that antibiotic use within a cohort of children with similar lifestyles predisposes towards atopy. Further insights into what is now referred to as the ‘gut-lung axis’ 41 describe how antibiotic treatment followed by oral administration of C. albicans renders immunocompetent hosts susceptible to allergic airways disease. Similarly, oral administrations of Mycobacterium vaccae,49helicobacter pylori50 as well as conventional probiotic strains51 have been shown to ameliorate symptoms of allergic airways disease in mice. Several of these studies highlight the necessity of neonatal administration rather than adult, emphasising that immune modulation is most effective during the developmental period.52
ProbioticsAnother crucial method of modulating the intestinal flora consists in administering live, viable bacteria via food or medicinal products, namely probiotics. This suggestion is based on the epidemiological data which have shown that atopic children have a different intestinal flora from that of healthy children, with higher levels of Clostridia and lower levels of Bfdba. Thus, probiotics are being tested currently for their efficacy in the prevention and therapy of allergy in infants.53,54
The targets of the probiotic therapy in food allergy need to be elucidated. Thus far, specific probiotics were shown to reverse the increased intestinal permeability,55 characteristic of children with atopic eczema and food allergy 56, and to enhance specific IgA responses,55,57,58 frequently defective in children with food allergy.59 Promotion of gut barrier functions by probiotics also includes normalisation of the gut microecology, alterations which have been demonstrated in allergic subjects 19 and generally modulation of mucosal and systemic immune responses.60 Importantly, the effects of probiotic bacteria are species and strain-specific, so it is imperative to select probiotic bacteria with specific activities based upon known in vitro or in vivo effects that are relevant to the clinical context they will be applied to. The various effects of different probiotic strains in food allergy are summarised in Table 2, according to randomised controlled trials, published in PubMed till 2012. The results of a metanalysis reviewed six studies enrolling 1549 infants and reported no other benefits of probiotics for food hypersensitivity.61 However, recent RCTs indicate that supplementation of EHCF (extensively hydrolysed casein formula) with LGG accelerated the development of tolerance in infants to cow's milk protein.62 Furthermore, it is of particular interest to state that probiotics have indeed been shown to exert distinct effects on antigen transport, depending on the food matrix, for example the quality of protein in the usual diet. In a rodent model, mucosal transport of degraded macromolecules has been found to be stimulated when Lactobacillus GG is administered together with unhydrolysed protein, but reduced when administered with hydrolysed protein.63 Such protein may thus stimulate the humoral immunity in the gut, but also affect the induction of oral tolerance, as antigen degradation is an indispensable component in the acquisition of mucosal tolerance.14
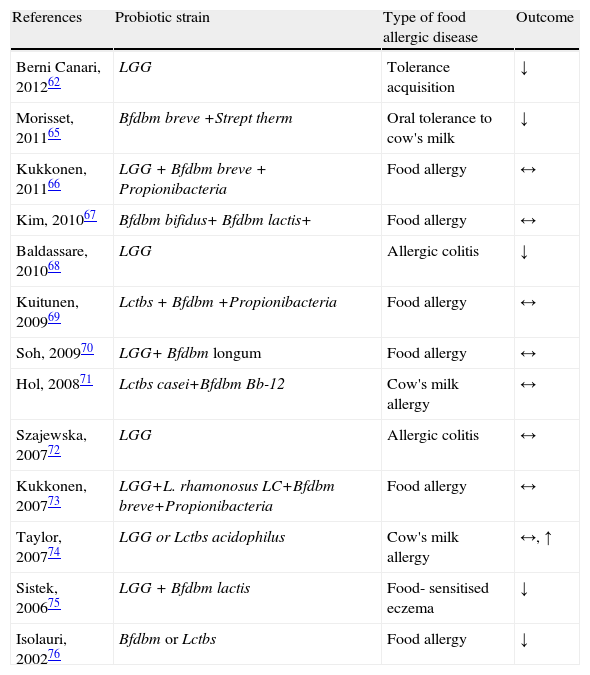
Various effects of different probiotic strains in mechanisms of food allergic disorders are shown from published randomised controlled trials.
| References | Probiotic strain | Type of food allergic disease | Outcome |
| Berni Canari, 201262 | LGG | Tolerance acquisition | ↓ |
| Morisset, 201165 | Bfdbm breve +Strept therm | Oral tolerance to cow's milk | ↓ |
| Kukkonen, 201166 | LGG + Bfdbm breve + Propionibacteria | Food allergy | ↔ |
| Kim, 201067 | Bfdbm bifidus+ Bfdbm lactis+ | Food allergy | ↔ |
| Baldassare, 201068 | LGG | Allergic colitis | ↓ |
| Kuitunen, 200969 | Lctbs + Bfdbm +Propionibacteria | Food allergy | ↔ |
| Soh, 200970 | LGG+ Bfdbm longum | Food allergy | ↔ |
| Hol, 200871 | Lctbs casei+Bfdbm Bb-12 | Cow's milk allergy | ↔ |
| Szajewska, 200772 | LGG | Allergic colitis | ↔ |
| Kukkonen, 200773 | LGG+L. rhamonosus LC+Bfdbm breve+Propionibacteria | Food allergy | ↔ |
| Taylor, 200774 | LGG or Lctbs acidophilus | Cow's milk allergy | ↔, ↑ |
| Sistek, 200675 | LGG + Bfdbm lactis | Food- sensitised eczema | ↓ |
| Isolauri, 200276 | Bfdbm or Lctbs | Food allergy | ↓ |
↑: Increase in symptoms or negative effect; ↓: decrease in symptoms or positive effect; ↔: no change in symptoms or no effect.
LGG, Lactobacillus rhamnosus GG; Lctbs acidophilus, Lactobacillus acidophilus; Lctbs casei, Lactobacillus casei; Bfdbm, Bifidobacterium; Bfdbm breve, Bifidobacterium breve; Strept therm, Streptococcus thermophilus; Propionibacteria, Propionibacterium freudenreichii spp.; Bfdbm lactis, Bifidobacterium lactis; Bfdbm bifidum, Bifidobacterium bifidum; Bfddm lactis, Bifidobacterium lactis.
In addition, the normal neonatal intestinal colonisation process may be markedly disturbed in premature infants, resulting in inappropriate colonisation and a predisposition to intestinal inflammation, such as the challenging clinical disease of necrotising enterocolitis.64 Theoretically, intervention that targets a positive modification in the intestinal flora could constitute an effective method of preventing the onset of this multifactorial entity,6 underscoring the importance of microbe-host interactions in an immune mediated disease.64
Correlation of the “hygiene hypothesis” with the “foetal programming” hypothesisBased on the hygiene hypothesis, in addition to environmental factors, the intestinal microflora is another contributor to food allergy by substantially affecting the mucosal immunity. Allergic responses are thought to arise if there is an absence of microbial exposure while the immune system is developing.77,78 The complex interactions between microbes and host combined with recent clinical observations and epidemiological trends may point to the convergence of two well-supported (although imperfect) hypotheses: the “hygiene hypothesis” and the “foetal programming hypothesis”. These specific interactions between the microbes and host during critical periods of immune development (including interactions between the foetus and the maternal immune system and microbiome during gestation) may have consequences well into adulthood.79
New technology will allow us to address the interactions of entire microbial communities and their role in immune development and function spanning from gestation through adulthood. Ultimately, these efforts will generate new models and interventions with the goal of promoting human health and preventing human disease.
New insights into gut microbiota connection with food allergyNew insights into gut microbiota connection with food allergy have arisen from experimental animal models. Evidence suggests that the microbial flora is a key environmental influence in programming oral tolerance. The lack of flora in germ-free (GF) mice is associated with the development of TH2 and IgE responses to food antigens.80 Microbial signals, such as those delivered by lipopolysaccharide (LPS) of the commensal bacterium Bacteroides fragilis or by a mix of clostridial species, induce mucosal tolerance by promoting the formation of induced regulatory T (iTreg) cells.81 Polymorphisms in or deficiency of genetic elements encoding microbial sensors, such as CD14, a high-affinity receptor for bacterial LPS, and Toll like receptor 4, which mediates responses to LPS, are associated with food allergy.82
Additionally, intestinal commensal bacteria and their sequential establishment play a crucial role in the development of gastrointestinal-associated lymphoid tissue and the modulation of the T-helper Th1/Th2/T regulatory balance, a major factor in the rise of food allergy.83 B-lactoglobulin (BLG)-stimulated splenocytes from sensitised conventional mice secreted significantly higher levels of IFN-γ and IL-10 and lower levels of IL-4 than splenocytes from germ-free counterparts. These data suggested that sensitised germ-free mice may have a Th2-skewed immune response and/or a defect in mounting a proper Th1/regulatory T-cell response.80 Furthermore, colonisation with staphylococci was correlated with less severity in the allergic response. It could therefore be linked to a protective effect against allergy or it may be an indicator of alterations in subdominant populations. 80
In a recent study,84 it was firstly demonstrated that a transplanted healthy infant microbiota mainly composed of Bifidobacterium and Bacteroides had a protective impact on sensitisation and cow's milk allergy in mice despite altered T-cell response in the ileum. The authors noted that the protective effect could be linked to a restored T-cell response over time in the ileum due to induced high ileal foxp3 gene expression, a master regulatory molecule in regulatory T-cells on allergies.85 The exact mechanisms of protection by regulatory T cells are unclear, but the release of suppressor cytokines and the direct suppression of target cells by cell contact remain the principal purported pathways.86
Concluding remarksIt is conceivable that food allergy is a complex and multifactorial disease, the clinical expression of which is determined by the interplay between host and environmental influences on gut microbiota during infancy. During this substantial period in human life, the specific formulation of gut microbiota might influence key immunological events that enhance allergic sensitisation to food. Regarding the maternal–foetal interaction, allergic mothers may transfer specific species of microbiota to their infants leading to aberrant compositional development of the microbiota, which may predispose infants towards allergic diseases. Furthermore, infants born vaginally in a hospital setting have a different GM composition. These alterations are associated with an increased risk of sensitisation to food allergens until the age 6–7 years old. In addition, breast fed infants appear to be in bifidobacteria colonisation in a range from 60% to 90%, which set the basis for gut microbiota development and modulation. Interestingly, changes in the microbiota populations can also be implemented by antibiotic therapy, resulting in a long-term decrease in beneficial anaerobic organisms and an increase in potentially harmful microbes. The accumulating evidence also indicates that probiotics supplementation effects are strain-specific and have great potential in the management of food allergy, related to their immunomodulatory effects.
Thus, the concept of vaginal home delivery and breast feeding is in line with minimising the risk for the development of food allergy sensitisation. Likewise, it is advisable to think seriously about prescribing long-term antibiotics for non-life threatening conditions and also consider probiotic strategies as an excellent tool with which to achieve controlled modification of the intestinal microflora.
Ethical disclosuresPatients’ data protectionConfidentiality of data. The authors declare that no patient data appears in this article.
Right to privacy and informed consentRight to privacy and informed consent. The authors declare that no patient data appears in this article.
Protection of human subjects and animals in researchProtection of human and animal subjects. The authors declare that no experiments were performed on humans or animals for this investigation.
Conflict of interestThe authors have no conflict of interest to declare.