Tuberculosis (TB) continues to be a serious worldwide public health problem today. According to the latest global data provided by the World Health Organisation (WHO), in 2016 there were approximately 10.4 million new cases and 1.7 million deaths worldwide from this infectious respiratory disease. Since the discovery of the first drugs, resistant strains began to appear, making it necessary to treat the disease with a combination of tuberculostatic drugs. Strains are considered to be multi-drug resistant (MDR) when resistance is detected to at least isoniazid (H) and rifampicin (R). When such resistance is not identified prior to the start of treatment, patients infected with mono-resistant strains have a high risk of developing additional resistance if they receive standard therapy.1
Nucleic acid amplification tests enable rapid identification of the M. tuberculosis complex (MTBc) in clinical samples. Xpert® MTB/RIF (Cepheid®) is a system capable of detecting MTBc and resistance to R within 2h, with a sensitivity of 98% in samples with positive microscopy and 67% with negative microscopy, and with a specificity in both cases of 99%.2 In 2011, the WHO recommended it as a first diagnostic step in countries with high rates of MDR-TB.3
We present the case of a patient diagnosed with pulmonary TB resistant to R according to Xpert® MTB/RIF (Cepheid®), but sensitive by the BD BACTEC™ MGIT™ 960 SIRE phenotypic method.
This was a 50-year-old male with a history of injecting drug use, active smoker, chronic alcoholism, and chronic hepatitis C.
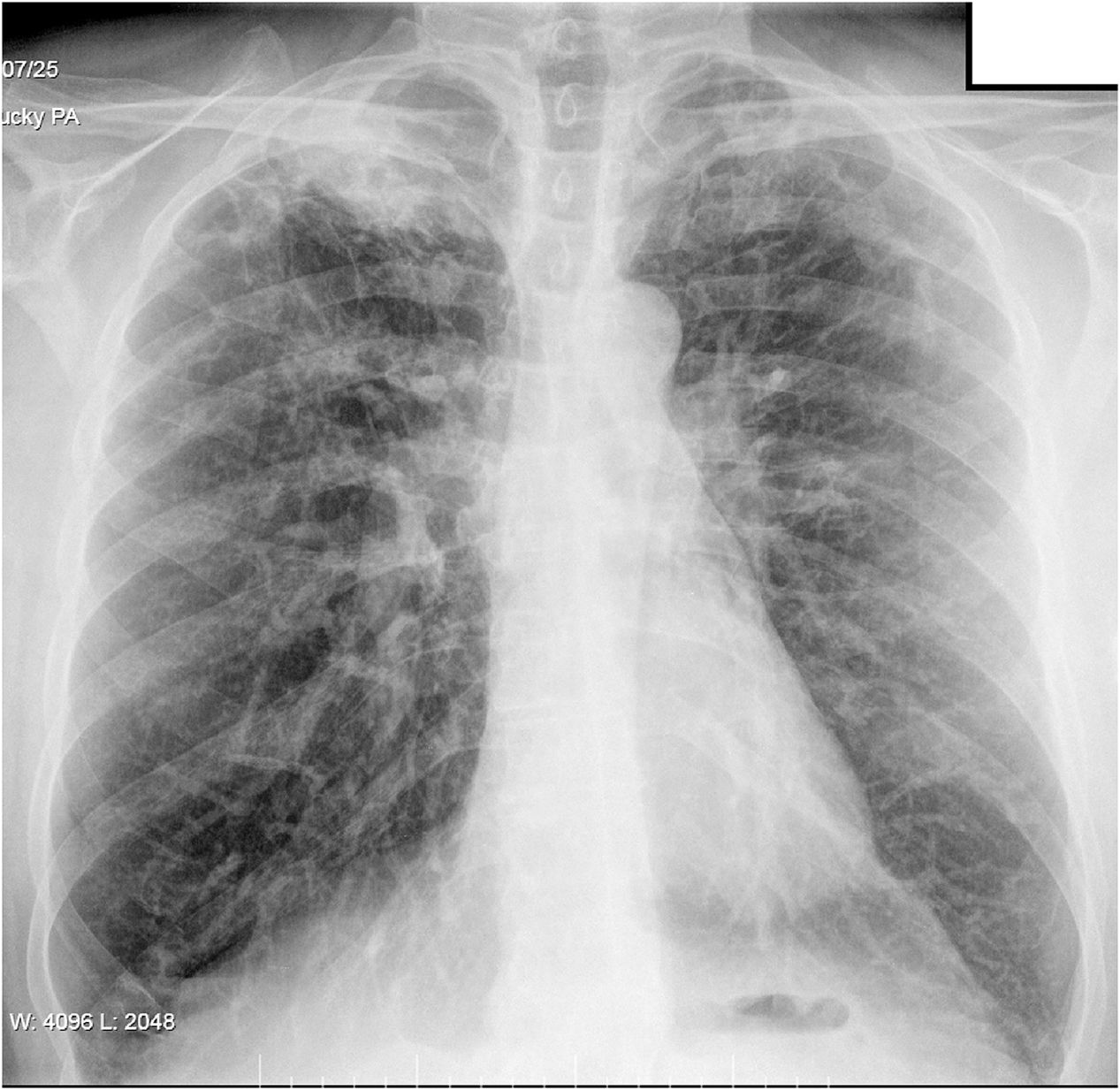
He reported having had a cough for eight months, which was initially dry, but with purulent sputum in the previous few days, although no haemoptysis. He had no pyrexia, constitutional syndrome or apparent contact with individuals with respiratory disease. Chest X-ray showed right apical infiltrate (Fig. 1) and sputum smear microscopy was positive.
Molecular detection of MTBc performed by Xpert® MTB/RIF (Cepheid®) was positive and resistance to R was identified. Culture in BD BACTEC™ MGIT™ 960 liquid medium detected growth after 19 days of incubation. The antibiogram performed by BD BACTEC™ MGIT™ 960 SIRE revealed sensitivity to H, R, streptomycin (S) and ethambutol (E). In view of the discrepancy, the strain was sent to the Mycobacteria Genetics Group at the University of Zaragoza for sequencing of the rpoB gene resistance determining region; the L511P mutation was detected, which explained the difference.
As the patient was initially diagnosed with pulmonary TB resistant to R, it was decided to start treatment with the following regimen: two months of quadruple treatment (H+Z+E+levofloxacin) and completing up to 12 months with the first three. Although it was later reported that the mutation that conferred genotypic, but not phenotypic, resistance to R was L511P, the same treatment pattern was maintained, with good progress and negative repeat culture after two months of treatment.
The treatment of TB consists of two phases: intensive and continuation. The aim of the continuation phase is to prevent relapses after treatment is completed. In this phase, R plays a key role, as it is the most effective drug because of its sterilising effect capable of eliminating the persistent bacilli responsible for recurrences. Isolated resistance to R therefore determines the prognosis of MDR-TB.4
According to both the Mensa and Sanford antimicrobial therapy guidelines from 2016, in cases of resistance to R, treatment with H, Z and E is recommended for 12 months. In the first two months, levofloxacin or S can be added in patients with extensive lesions.5,6 However, according to the latest WHO recommendations, the patients should be treated as patients with MDR-TB. Conventional treatment would consist of a regimen of at least five drugs effective during the initial phase (eight months with Z and four second-line drugs), with a total duration of approximately 20–21 months. Because of the long duration, poor tolerance and toxicity, this regimen has not achieved success rates any higher than 55–70%. As a result, the WHO promoted a shorter nine-month regimen (Bangladesh regimen) consisting of a four-month intensive phase (capreomycin/amikacin+moxifloxacin+prothionamide/ethionamide+clofazimine+E+Z+H) and five-month continuation phase (moxifloxacin+clofazimine+E+Z) in patients who had not received fluoroquinolones or second-line drugs by injection, or when in vitro sensitivity to these antibiotics was demonstrated.7
Isolated resistance to R is very rare and is caused in more than 96% of cases by mutations in an 81bp region of the rpoB gene (RRDR, codons 507 to 533 referred to E. coli) which encode the β subunit of the RNA polymerase. The most common mutations are: H526Y and S531L. Mutations have also been found in phenotypically sensitive strains, such as 510H, D516Y, N518D, H526N and L533P, including the one detected by us, L511P.8
In the study by Ocheretina et al., in seven cases with discrepancy in resistance to R (phenotypic sensitivity and genotypic resistance) the mutation detected was L511P, and they were considered as isolates with low level of resistance to R (MIC>0.031μg/ml). In all the strains, the L511P mutation was associated with the S315T mutation in katG, which conferred additional resistance to H. In two of the cases, the L511P mutation was associated with M515T, which increased the MIC of R to 0.25–0.5μg/ml. Three of the patients with strains carrying the L511P mutation had therapeutic failure with both conventional and alternative regimens.9
In 2016, Gonzalo et al. detected three strains of sensitive MTBc by BD BACTEC™ MGIT™ 960 SIRE and carriers of the L511P mutation associated with M515I, whose MIC of R determined by microdilution were in the range of 4–8μg/ml. In this study, no clinical data were collected, so it was not possible to find out the clinical outcomes of patients carrying MTBc strains resistant to R.10
As suggested by the aforementioned authors, and based on the work of Gumbo et al., it would therefore be interesting to redefine the critical concentration of R, established in 1963 at 1μg/ml. The current approach to defining cut-off points uses population models based on pharmacokinetic and pharmacodynamic (PK/PD) principles. This could decrease the critical concentration of R even to 0.0625μg/ml,11 increasing the specificity of molecular tests and helping to differentiate between hidden cases of low-level resistance and silent mutations.
We conclude that the clinical significance of mutations, such as the L511P we detected, needs to be assessed, as the discrepancy between methods such as Xpert® MTB/RIF and the gold standard BD BACTEC™ MGIT™ 960 SIRE in the detection of resistance to R is going to become an increasingly common finding in clinical laboratories.
Conflicts of interestThe authors declare that they have no conflicts of interest.
To Dr Sofía Samper Blasco for the critical review of this manuscript and all the members of the Mycobacteria Genetics Group at the University of Zaragoza for their contribution in the molecular analysis of the MTBc strain under study.
Please cite this article as: Rapún Mas L, Angulo López I, Cecilio Irazola Á, Betrán Escartín AI. Discrepancia en la resistencia genotípica versus fenotípica a rifampicina en Mycobacterium tuberculosis. A propósito de un caso. Enferm Infecc Microbiol Clin. 2019;37:212–213.