Candida parapsilosis sensu stricto, Candida orthopsilosis, and Candida metapsilosis are cryptic species that belong to the C. parapsilosis complex, which has been increasingly associated to fungemia in various geographic regions, principally due to the capability of these yeasts to form biofilms on indwelling medical devices. BCR1 is one of the most studied genes related to Candida spp. biofilms.
AimsTo evaluate the biofilm forming capability of a subset of 65 clinical isolates of the C. parapsilosis complex using two conventional approaches, and to look for an association between the biofilm forming phenotype and genetic variants of a fragment of BCR1.
MethodsThe biofilm determination was carried out by crystal violet staining and tetrazolium reduction assay. On the other hand, a segment of BCR1 gene was sequenced by Sanger methodology.
ResultsC. parapsilosis sensu stricto was statistically associated with a low biofilm production phenotype, while C. orthopsilosis was significantly associated with both phenotypes (high and low biofilm producers). According to the BCR1 sequence analysis, genetic variability was detected in C. orthopsilosis and C. metapsilosis without a particular biofilm formation phenotype association.
ConclusionsUnder the adopted experimental design, C. parapsilosis sensu stricto was associated with the low biofilm phenotype and C. orthopsilosis with both phenotypes (high and low biofilm producers). On the other hand, an association between a biofilm forming phenotype and a particular genetic variant of the analyzed BCR1 fragment was not found.
Candida parapsilosis sensu stricto, Candida orthopsilosis y Candida metapsilosis son especies crípticas que integran el complejo C. parapsilosis, asociado de forma creciente a fungemia en diversas regiones geográficas. Dicho crecimiento se debe principalmente a la capacidad de estas levaduras de crear biopelículas en los dispositivos médicos. El gen BCR1 es uno de los más estudiados en las biopelículas de Candida spp.
ObjetivosEvaluar la capacidad de formación de biopelícula de un conjunto de 65 aislamientos clínicos del complejo C. parapsilosis mediante dos metodologías convencionales, así como establecer una posible asociación entre el fenotipo productor de la biopelícula y las variantes genéticas de un fragmento de BCR1.
MétodosLa determinación de la presencia de biopelícula se llevó a cabo mediante tinción con cristal violeta y el análisis de reducción de la sal de tetrazolio. Además, se secuenció un segmento del gen BCR1 mediante el método Sanger.
ResultadosC. parapsilosis sensu stricto presentó una asociación estadísticamente significativa con un fenotipo de baja producción de biopelícula, mientras que C. orthopsilosis tuvo una asociación estadísticamente significativa con ambos fenotipos (alta y baja producción de biopelícula). Según el análisis de la secuencia de BCR1, existe variabilidad genética en C. orthopsilosis y C. metapsilosis sin ninguna asociación particular a los fenotipos relacionados con la formación de biopelícula.
ConclusionesBajo el diseño experimental adoptado, C. parapsilosis sensu stricto se asoció con un fenotipo de baja producción de biopelícula y C. orthopsilosis con ambos fenotipos (alta y baja producción de biopelícula). Por otra parte, no se encontró ninguna asociación estadísticamente significativa entre los fenotipos de formación de biopelícula y variantes genéticas particulares en el fragmento analizado de BCR1.
The Candida parapsilosis complex (C. parapsilosis sensu stricto, C. orthopsilosis, and C. metapsilosis) currently stands out as the second or third most common yeast species isolated from blood cultures in Latin America, Canada, Europe and Asia,1,9,15 becoming an important focus of attention, particularly among neonates and immunocompromised patients.31 These opportunistic pathogens have been recovered from the hands of health-care workers,1 and are well known as a cause of bloodstream infections associated with parenteral hyperalimentation and intravascular devices due to their capability to form biofilms.21
Biofilms are complex surface-associated microbial communities embedded within an extracellular matrix. They are considered the most prevalent growth form of microorganisms.24 Biofilm formation is an important virulence factor for a variety of Candida species, including C. parapsilosis, as it confers significant tolerance to antifungals and protects yeasts cells from host immune responses,25 whereby biofilms themselves are a reservoir for persistent infections.2
BCR1 (Biofilm and Cell wall Regulator 1) is a conserved fungal transcription factor required for biofilm formation in both Candida albicans and C. parapsilosis.4,16,18 Some major targets of Bcr1 in C. albicans include genes coding for adhesins and cell-wall proteins, suggesting that Bcr1 is involved in the early adhesion stage of biofilm development.5,16,18 However, it was demonstrated that the biofilm formation process in C. parapsilosis could be dependent and independent of BCR1.20
The aim of this study was to quantify biofilm formation of a subset of 65 clinical isolates of the C. parapsilosis complex by two different methodologies: crystal violet staining and tetrazolium (XTT) reduction assay, as well as to analyze the nucleotide sequence of a fragment of the BCR1 gene looking for a possible association between the biofilm forming phenotype and genetic variants of the gene segment analyzed.
Materials and methodsIdentification of isolatesA total of 65 clinical isolates of C. parapsilosis from a previous work30 were included in this study. These were grown at 37°C for 24h on Potato-dextrose agar (PDA) (Difco, USA). The isolates were initially identified as C. parapsilosis by API 20C AUX strips (bioMérieux, Mexico) and standard morphological methods. Re-identification of the isolates was then performed using RFLP-BanI.28 The isolate study collection consisted of 29 isolates each of C. parapsilosis sensu stricto and C. orthopsilosis, and 4 isolates of C. metapsilosis. A blood origin accounted for 48% of the isolates, while the remaining 52% were as follows: nail (9 isolates), skin (8 isolates), peritoneal fluid (7 isolates), urine (3 isolates), bronchial fluid (2 isolates), and 1 isolate each from spinal fluid, bile and otic secretion. Type strains C. parapsilosis sensu stricto ATCC 22019, C. orthopsilosis ATCC 96139, and C. metapsilosis ATCC 96144 were used as quality controls. All isolates were stored as suspensions in sterile distilled water at room temperature until testing.
Biofilm induction and quantitationThe growth conditions and biofilm formation evaluated by crystal violet staining and XTT reduction assays were done according to the methodology described by Melo et al.14. Strains were initially grown on Sabouraud-dextrose agar (SDA) (Difco, Detroit, MI, USA) at 37°C for 24h, and one colony of each strain was further subcultured in RPMI 1640 broth medium with l-glutamine (Hardy Diagnostics, Santa Maria, CA, USA) at 37°C for 18h at 120–200rpm. The cells were harvested, washed twice with PBS, and adjusted to a concentration of 1×107cells/ml in RPMI 1640 medium. An aliquot of 100μl of the cell suspensions were transferred into each well of a sterile flat-bottomed 96-well polystyrene Corning Costar 9018 plate (Costar, Corning Incorporated, NY, USA). The plates were incubated at 37°C for 1.5h at 75rpm, washed with 150μl of PBS and then 100μl of fresh RPMI 1640 medium was added to each well. The plates were finally incubated at 37°C for 72h at 75rpm to allow biofilm growth. Test medium without cells was added to the final well of each plate as the negative control.
To determine bulk biofilm formation, the crystal violet staining assay was performed. Briefly, after biofilm formation, each well was washed twice with 200μl of PBS and the plates were dried at 37°C for 20min. The washed biofilms were stained with 110μl of 0.4% aqueous crystal violet solution for 45min. The wells were then washed three times with 200μl of sterile distilled water and then faded with 200μl of 95% ethanol. After 45min, 100μl of detaining solution from each sample was transferred to a new plate and measured with a spectrophotometer plate reader at 595nm. The optical density (OD) values of the negative controls were subtracted from the values of the test wells in order to eliminate background interference. On the other hand, biofilm metabolic activity was determined by the XTT (tetrazolium salt 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-5-(phenylamino)-carbonyl-2H-tetrazoliumhydroxide) (Sigma, USA) reduction assay. After biofilm formation, the plate wells were washed twice with 200μl of PBS, then 200μl of PBS and 12μl of XTT-menadione solution were added to each well. The XTT-menadione solution was prepared on each day of testing by adding 1.5ml of XTT (1mg/ml in sterile saline) to 300μl of menadione solution (0.4mM in acetone; Sigma, St Louis, MO, USA). Plates were incubated at 37°C in darkness for 5h. Afterwards, 100μl of the reaction solution was transferred to a new plate and the concentration of the formazan product was spectrophotometrically determined at 490nm. The OD values of the negative control wells were subtracted from the values of the test wells. The amount of biofilm formed by an isolate was categorized as high (OD490≥0.1), low (0.025≤OD490<0.1) and no biofilm formation (OD490<0.025), according to the criteria previously established by Pannanusorn et al.19
BCR1 gene amplification and sequencingYeast genomic DNA of each clinical isolate was extracted from a homogenized pellet using the phenol-chloroform method,23 and then resuspended in nuclease-free water. Total genomic DNA was treated with RNase I (Invitrogen, USA) for 30min at 37°C to remove RNA traces, and the quality and integrity were assessed by standard spectrophotometric and elecrophoretic methods, respectively. Due to the fact that BCR1 gene sequence or trace archives of C. metapsilosis were not available at sequence data bases at the moment the experiments were done, the design of a consensus primer set to amplify BCR1 in C. parapsilosis complex (Sense 5′-AAA ACA CAC YGG TGA ACG AC-3′ and Anti-sense 5′-TCA CTC ATK GCT GAT CTT GG-3′) was based on the available highly conserved sequences previously reported for C. parapsilosis strain CDC317 (accession number: HE605206) and C. orthopsilosis strain Co 90-125 (accession number: HE681722) at GenBank (www.ncbi.nlm.nih.gov) using the online primer-3 tool (http://biotools.umassmed.edu/bioapps/primer3_www.cgi). This primer set was used at 5μM with the commercial kit PCR master mix of Promega (USA). The amplification reactions were designed in a final volume of 25μl and carried out in a Veriti 96-Well Thermal Cycler (Applied Biosystems, Thermo Fisher Scientific Inc., Waltham, MA). The standardized cycling parameters were as follows: 1 initial denaturation step of 4min at 94°C, 42 cycles of 1min at 94°C, 30s at 57°C, 90s at 72°C, and finally an elongation step of 6min at 72°C. The PCR products were visualized on 2% agarose gels stained with ethidium bromide and visualized under UV light (UVP BioImaging Systems, EpiChemi 3 Darkroom UVP Inc., Upland, CA). Amplicons were sequenced using Big Dye terminator cycle sequencing kit v3.1 using the same PCR primers. The reactions were analyzed in the ABI PRISM 3100 Genetic Analyzer using the Sequencing Analysis Software v5.3 of Applied Biosystems. The obtained sequences of each isolate were aligned using the CLUSTAL-W program and GeneStudio Pro software (GeneStudio, Inc., Suwanee, GA, USA), followed by manual corrections if needed. Sequences were then annotated using Sequin software from NCBI and the nucleotidic sequences were finally deposited in GenBank under the following accession numbers: KJ610843 to KJ610856.
StatisticsThe biofilm quantifications were carried out twice at different times, with two technical replicates each time. The OD values were expressed as means±standard deviation (SD) for each set of data determined and were compared using Student's t test (SPSS v17.0 for Windows; SPSS Inc., Chicago, IL). The biofilm categorization as high, low and negative biofilm production was compared among the C. parapsilosis species complex, clinical origin and genetic variants of BCR1 (looking for a possible association) by Pearson Chi-squared test; a P-value ≤0.05 was considered significant. The graphics were performed on GraphPad Prism v5.03 for Windows (GraphPad Software, San Diego, CA).
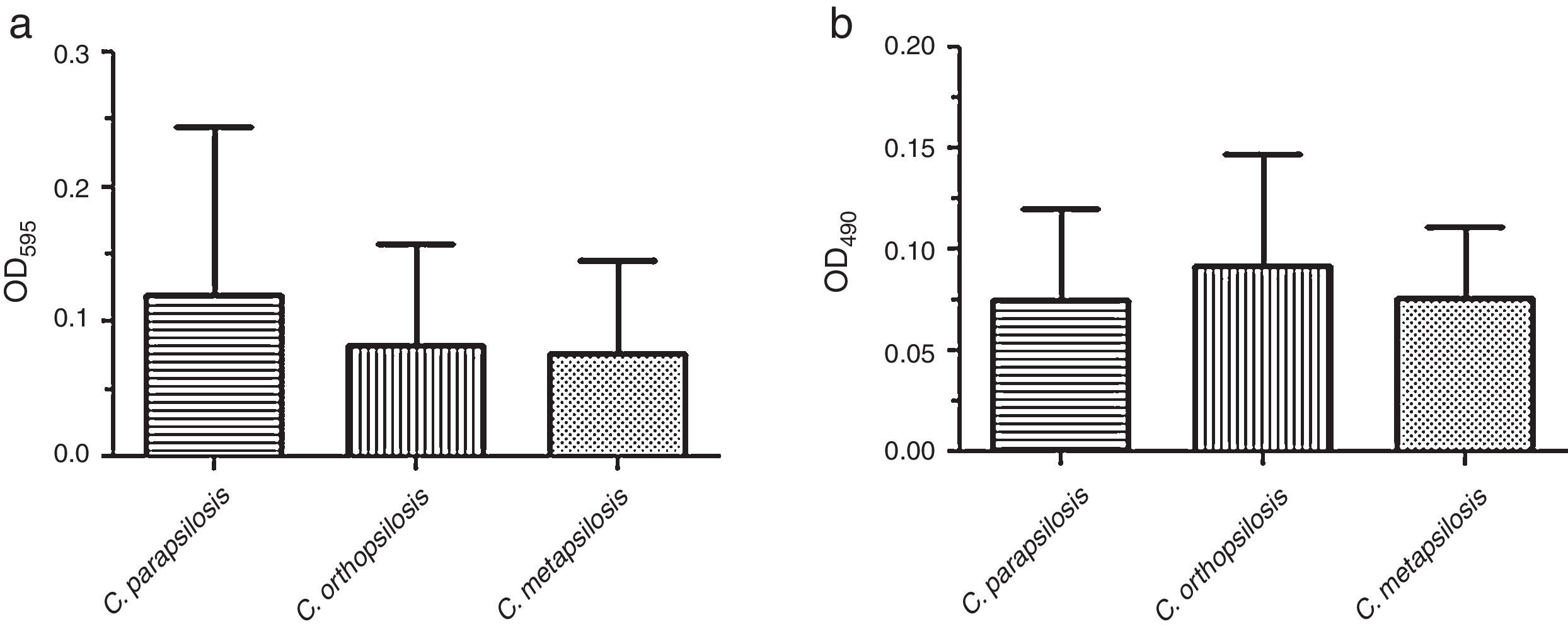
ResultsBiofilm quantification by crystal violet staining is showed in Fig. 1(a). In this assay, the mean OD595 value and range for each species were as follows: C. parapsilosis sensu stricto, 0.121 (SD+0.124) with a range of 0.014–0.571; C. orthopsilosis, 0.082 (SD+0.076) with a range of 0.006–0.293; C. metapsilosis, 0.077 (SD+0.068) with a range of 0.013–0.175. C. parapsilosis sensu stricto showed the highest biofilm production as determined through crystal violet staining.
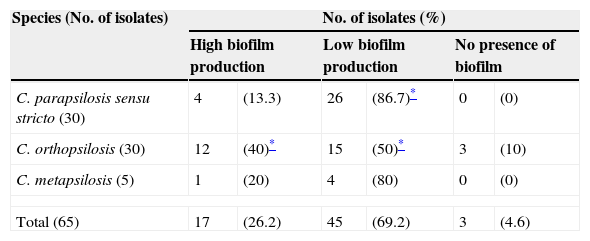
Fig. 1(b) depicts the biofilm metabolic activity of the tested isolates. In this assay, the mean OD490 value and range for each species were as follows: C. parapsilosis sensu stricto, 0.075 (SD+0.045) with a range of 0.028–0.247; C. orthopsilosis, 0.092 (SD+0.054) with a range of 0.014–0.235; C. metapsilosis, 0.076 (SD+0.035) with a range of 0.049–0.135. C. orthopsilosis showed the highest biofilm production as determined through XTT reduction assay. Additionally, according to Pannanusorn et al. criteria,19 62 isolates (95.4%) produced biofilm, 26.2% were high biofilm producers, while 69.2% were categorized as low biofilm producers. Biofilms were predominantly highly produced by C. orthopsilosis (40%), whilst C. parapsilosis sensu stricto was mostly a low biofilm producer (86.7%). The non-biofilm producer phenotype was detected in just 3 isolates of C. orthopsilosis (4.6%) (Table 1). C. parapsilosis sensu stricto was statistically associated with low biofilm production (P=0.0108), while C. orthopsilosis was significantly associated with both high and low biofilm production (P=0.0386 and P=0.0045, respectively).
Categorization of biofilm formation of clinical isolates pertaining to C. parapsilosis complex, assessed by metabolic activity determination.
All the isolated DNAs were under the quality and quantity standards for PCR reaction. The designed consensus primer set amplified a fragment of BCR1 gene. These amplicons included a segment of exon and intron. From all the isolates a PCR product was generated. The nucleotide sequence analysis of amplicons corresponded to a fragment of the BCR1 gene previously reported in the BLAST analysis. These sequences showed genetic variability among the analyzed species. A total of eight genetic variants (designed as: Co-1 to 8) ranging from 913 to 1025bp, were detected in C. orthopsilosis, while C. metapsilosis presented two distinct genotypes of 872bp (Cm-1) and 984bp (Cm-2); moreover, genetic variability was not detected in C. parapsilosis sensu stricto, which amplified an approximate 872 bp-product (Cp-1).
In the obtained sequences, we located the section corresponding to the exon, which was aligned. The open reading frame (ORF) was deducted from it, and also this sequence (supplementary material, Fig. S1) was aligned. According to the fragment of BCR1 exon analysis, the sequence of C. parapsilosis ATCC 22019 was identical to all C. parapsilosis sensu stricto strains (Cp-1). Moreover, the sequence of C. metapsilosis ATCC 96144 was the same to Cm-2, and 68.14% similar with respect to Cm-1, which is equal to the C. parapsilosis sensu stricto strains. On the other hand, the fragment of BCR1 exon of C. orthopsilosis ATCC 96139 was identical to those of C. parapsilosis sensu stricto strains, and conserved a homology ranging from 60.14% to 79.45% with respect to the eight genetic variants of C. orthopsilosis (Co-1 to 8). Finally, the most frequent variant of the analyzed fragment of BCR1 exon in C. parapsilosis complex, with 49.2% (32 isolates) was shared by C. parapsilosis ATCC 22019, Cp-1, C. orthopsilosis ATCC 96139, and Cm-1, which at the same time is identical to the deposited sequence of C. parapsilosis strain CDC317 (GenBank accession number: HE605206). Finally, statistical associations between variants of the analyzed segment of BCR1 gene exon and the biofilm forming phenotypes of the isolates were analyzed by Pearson Chi-squared test, but were not found. Different genetic variants of BCR1 were classified within more than one biofilm forming phenotype, without any evident pattern of association with statistical support.
DiscussionThe capability of Candida spp. to form biofilms may confer an ecological advantage to yeasts, aiding their survival as human commensals and pathogens. This may also be responsible for making them particularly well adapted to colonization of host tissues and indwelling devices.24 To date, several methods for assessing Candida biofilm production have been reported.3 In the present study, we evaluated the biofilm forming capability of a subset of 65 clinical isolates of the C. parapsilosis complex by two different techniques, crystal violet staining and XTT reduction assay. Crystal violet staining is probably the most reliable assay for determining bulk biofilm formation because it considers both metabolically-active and inactive cells of biofilms.22 Another method that has been widely used to quantify Candida biofilms in vitro is XTT reduction assay,11 which specifically considers the biofilm metabolically-active cells through XTT reduction by mitochondrial dehydrogenases.
The study of biofilm forming capability among the C. parapsilosis species complex have not yet been clearly understood due to apparently contradictory findings. Song et al.26 studied the capability of 159 isolates of the C. parapsilosis complex to produce biofilm through a spectrophotometric approach in which cultivated biofilms were incubated without agitation, and transmittance values were measured without previous staining at 405nm. They reported that C. parapsilosis sensu stricto (formerly C. parapsilosis group I) was the only species of the complex able to form biofilms. Later, Tavanti et al.29 using the same methodology previously mentioned with few modifications, reported that none of the 33 C. orthopsilosis clinical isolates they studied could form biofilms in vitro. In contrast, Lattif et al.13 based on XTT reduction assay, dry weight measurement, scanning electron microscopy and confocal laser scanning microscopy of 10 clinical isolates each of C. parapsilosis sensu stricto, C. orthopsilosis and C. metapsilosis, established that all three species were able to form biofilms, which were similar in surface topography and architecture. Finally, Melo et al.14 confirmed the biofilm forming capability of the C. parapsilosis species complex through the analysis of 20 isolates (7 C. parapsilosis sensu stricto, 8 C. orthopsilosis and 5 C. metapsilosis) by crystal violet staining, XTT reduction assay and scanning electron microscopy.
The results we obtained about the biofilm forming capability of all three C. parapsilosis species complex on polystyrene microtiter plates under our experimental conditions are in contrast to those reported by Song et al.26 and Tavanti et al.29, and in agreement with those reported earlier by Lattif et al.13 and Melo et al.14. Although P values were not significant, according to crystal violet staining, the rank scale for biofilm production was C. parapsilosis sensu stricto>C. orthopsilosis>C. metapsilosis, while based on XTT reduction assay the rank was C. orthopsilosis>C. metapsilosis≥C. parapsilosis sensu stricto. Otherwise, the origin and selection of isolates can be one factor that influences biofilm formation in Candida spp.8,10 Recent reports have demonstrated that blood isolates produce greater quantities of biofilm compared with other clinical origins.12 However, we have not found a significant statistical difference between the biofilm forming capability and the origin of the tested isolates, agreeing with Silva et al.24
The microbial biofilm formation is a complex biological process finely regulated by inherent genetic mechanisms of the participating organisms. One of the master regulators of this phenomenon is the zinc finger transcription factor Bcr1.6 It was identified in screens for mutant strains defective in biofilm formation on abiotic surfaces16,17 and in adherence to a silicone substrate.7 Furthermore, Bcr1 is required for expression of several cell surface protein genes, such as ALS1, ALS3, and HWP1, which are the major functional Bcr1 targets.16,17 Recently, Srikantha et al.27 demonstrated that BCR1 gene conferred impermeability, impenetrability, and drug resistance to a/α biofilms of C. albicans. Particularly in C. parapsilosis, biofilm formation is both dependent and independent of BCR1, but even in strains which showed a BCR1 independent biofilm phenotype BCR1 has alternative physiological functions.20 We did not find any association between a particular genetic variant of the analyzed BCR1 fragment and a biofilm forming phenotype of the C. parapsilosis complex isolates studied.
Conflict of interestThe authors have no conflicts of interest to declare
We thank Sergio Lozano-Rodriguez, M.D. for the English review of the manuscript prior to submission.