Human tuberculosis is still a major world health concern. In Uruguay, contrary to the world trend, an increase in cases has been observed since 2006. Although the incidence of MDR-resistant strains is low and no cases of XDR-TB were registered, an increase in the number of patients with severe tuberculosis requiring critical care admission was observed. As a first aim, we performed the analysis of the genetic structure of strains isolated from patients with severe tuberculosis admitted to an intensive care unit. We compared these results with those corresponding to the general population observing a statistically significant increase in the Haarlem genotypes among ICU patients (53.3% vs 34.7%; p<0.05). In addition, we investigated the association of clinical outcomes with the genotype observing a major incidence of hepatic dysfunctions among patients infected with the Haarlem strain (p<0.05). The cohort presented is one of the largest studied series of critically ill patients with tuberculosis.
La tuberculosis (TB) aún representa un problema mayor de salud pública. En Uruguay, contrariamente a la tendencia mundial, se ha observado un incremento en el número de casos desde 2006. Aunque la incidencia de casos de multidrogorresistencia (MDR) es baja y no se han reportados casos de resistencia a fármacos de primera y segunda línea de tratamiento (XDR), se ha observado un incremento en el número de casos con TB grave, que requieren internación en unidad de terapia intensiva (CTI). Como primer objetivo del presente trabajo, se analizó la estructura genética de cepas de Mycobacteriumtuberculosis aisladas de pacientes internados en CTI. Comparamos estos resultados con los obtenidos con cepas circulantes en la comunidad. Observamos un incremento estadísticamente significativo del genotipo Haarlem en los pacientes internados en CTI (53,3 vs. 34,7%; p<0,05). Además, investigamos la asociación del desenlace clínico con el genotipo, y encontramos una mayor incidencia de disfunción hepática en los pacientes infectados con la cepa Haarlem (p<0,05). La cohorte presentada en este trabajo corresponde a una de las series con mayor número de pacientes con tuberculosis que requirieron internación en CTI.
Although tuberculosis (TB) incidence is decreasing worldwide15, Uruguay has been reporting an increase in the number of cases since 2006. Despite the efforts made by the National Program Against Tuberculosis, the incidence rate in 2019 was 30 cases per 100000 inhabitants, placing Uruguay above the average of the region15. Based on this scenario, more cases with severe TB were diagnosed and admitted to Intensive Care Unit (ICU) during the last years.
TB in critical care is associated with respiratory and other organ failures and requires ICU admission for advanced vital support. Despite the availability of effective therapies, mortality rates remain between 15.5 and 65.9% in critical care patients10,11. In Uruguay, a high mortality rate (>50%) was observed in TB cases that required ICU admission.
Information from classical and molecular epidemiology, along with patient clinical data represent key elements to achieve better control of the disease3.
The genotypic variations, especially bacterial lineages, are associated with changes in transmission capacity and also in the ability to acquire drug resistance2,7–9. Thus, changes in the prevalence of specific lineages or sub-lineages could have important consequences for the observed epidemiology and outcome of TB disease12,14.
We hypothesized that critically ill TB patients could be associated with strain virulence. This study aimed to describe the genetic diversity of Mycobacteriumtuberculosis using classical genotyping methods of clinical isolates obtained from TB patients in the critical care setting, and to analyze them with the genetic background structure observed in community TB. In addition, we aimed to determine the possible association of comorbidities and organ dysfunctions observed in these patients according to the TB genotype causing the disease.
The research protocol used in this work was presented and approved by the Clinical Research Ethics Committee of Hospital Español, and has been performed following the ethical standards of the 1964 Declaration of Helsinki.
Sixty M. tuberculosis isolates obtained from critically ill patients from a general public hospital (Hospital Español – ASSE, Montevideo, Uruguay) were genotyped and compared to ninety-five randomly selected community isolates within the same period (Suppl. Table 1 and Suppl. Fig. 1).
All the community sample isolates correspond to patients treated with the standard drug protocol (daily HREZ for the first two months, four months HR 3 days a week). The cultures became negative after 3 months of treatment (Suppl. Table 1 and Suppl. Fig. 1). All the community samples were obtained from TB patients that did not require ICU admission at any time during treatment.
Patients with TB admitted to the ICU were classified according to a TB clinical severity score as having low, moderate, and high risk TB disease1. Comorbidities and organ dysfunctions were documented upon admission, along with common clinical and laboratory studies.
Isolate characteristics were summarized for each group (Community and Critically ill patient isolates) in Supplementary Table 1. In addition, the analysis pipeline was shown in Supplementary Figure 1. Microbiological cultures and DNA extractions were performed and stored, as previously described4. Molecular typing using spoligotyping and MIRU-VNTR were performed as described in Supplementary Figure 1.
Lineage assignment was carried out based on similarity search and UPGMA tree when comparing reference strains located in the server with our isolates with a distance less than 0.3 using www.miru-vntr.org tool. Comparative analysis between strain distribution of critical and non-critically ill patients was performed with the Chi-squared contingency tables for categorical data (p-value≤0.05 was considered a statistically significant difference). Minimum spanning trees were constructed for all isolates using the MST tool (miru-vntr.org).
Based on the documented risk factors, forty-five of our initial set of sixty-five patients (69%) could be classified according to TB severity at the moment of ICU admission into three groups (low, moderate and high severity). Most ICU admissions correspond to moderate to high severity TB (86.6%): low (13.4%), moderate (40.0%), and high risk (46.6%) (Suppl. Fig. 1)1.
Supplementary Figure 1 shows the main characteristics of TB patients admitted to an ICU including demographic data, bilateral pulmonary compromise, comorbidities, organ dysfunctions, relevant laboratory findings, classification according to a tuberculosis severity score1, and ICU mortality. In brief, patients were relatively young with male predominance. Bilateral pulmonary involvement was present in 44%, and HIV coinfection in 22% of the cases. Poor nutritional status was present in 76%. Respiratory and cardiovascular failure were the most frequent organ failures, and mechanical ventilation was implemented in 74% of the patients. Only 13.3% had a low tuberculosis risk severity score, while a moderate to high score was observed in 86.6% of these patients. Global mortality in ICU was 62% considering all patients (n=60) and reached 71.8% considering only high-to-moderate severity TB patients (n=39).
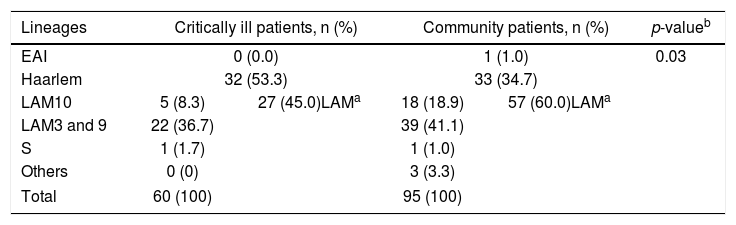
Among the ICU M. tuberculosis isolates, strains clustered into five main sub-lineages (Table 1A). Except for one particular case, all sub-lineages belong to Lineage 4 of M. tuberculosis. Typing of critical care patients reveals the Haarlem family as the most prominent in these samples (53.3%), followed by the LAM family (45.0%). On the contrary, LAM strains are more represented among community TB isolates (60.0%), followed by the Haarlem strains (34.7%), a significant statistical difference being observed when comparing these two populations (p<0.05).
Mycobacterium tuberculosis lineages in critically ill and community patients.
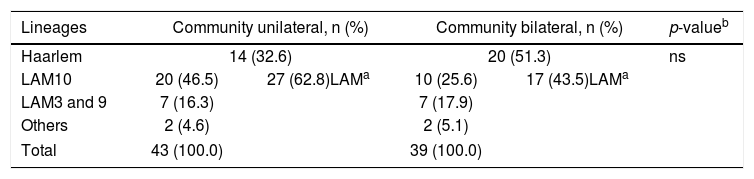
It is interesting to observe that the Haarlem family was more frequent in patients with bilateral TB (51.3%) versus unilateral TB (32.6%) in community patients (Table 1B). Although the observed difference was not statistically significant, it could indicate a more aggressive form of the disease caused by the Haarlem lineage. This hypothesis needs to be corroborated and could explain the high representation of the Haarlem genotype among critically ill patients.
Mycobacterium tuberculosis lineage comparison in pulmonary community patients with unilateral or bilateral TB compromise.
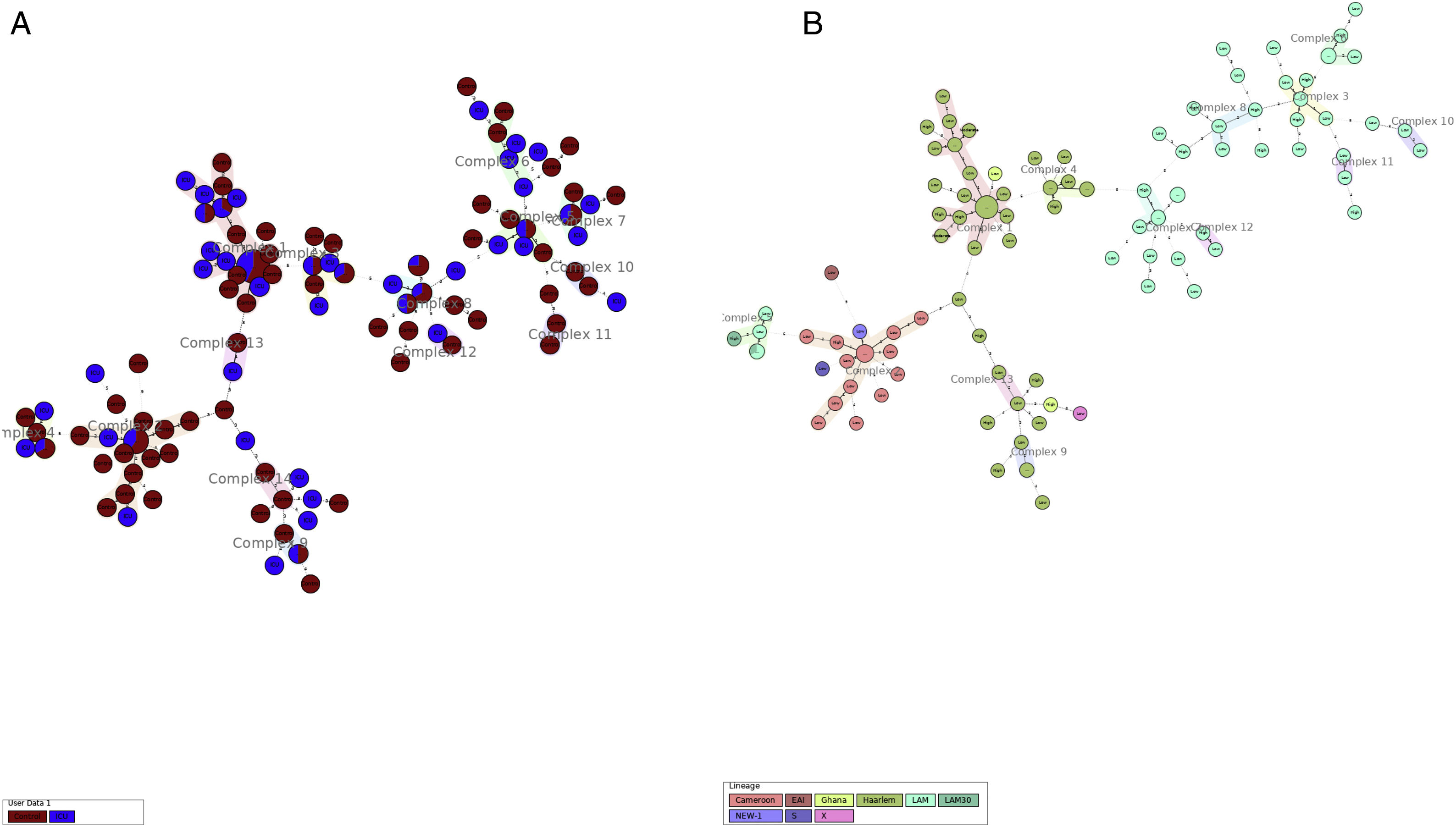
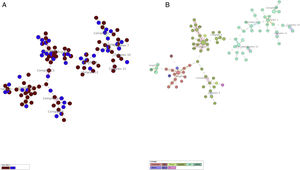
More detailed analyses of typing results were performed analyzing the minimum spanning tree generated with the online tool MST (miru-vntrplus.org). Fourteen clonal complexes were obtained, community and critical isolates could be detected in each cluster (Fig. 1A and Supplementary Table 2). The biggest cluster comprises 44 isolates (22 of them – 9 ICU and 13 community samples – with the same MIRU pattern belonging to the Haarlem genotype (Fig. 1B)). Finally, 24 community samples and 10 ICU samples remain unclustered.
Minimum spanning tree cluster analysis. (A) Samples in red correspond to ICU samples; samples in blue correspond to community samples. Samples of both groups were present in all clusters. (B) Same analysis colored by lineage. LAM and Haarlem lineages correspond to the most frequent isolates.
When analyzing the clonal complexes with more than six isolates (n=8), we could observe strains isolated during the whole studied period (Suppl. Table 2). This phenomenon also occurs when analyzing at group level. No clonal complexes were associated with a particular isolation year that could be due to a particular outbreak.
We investigated the possible association of observed TB genotypes with ICU patient's comorbidities.
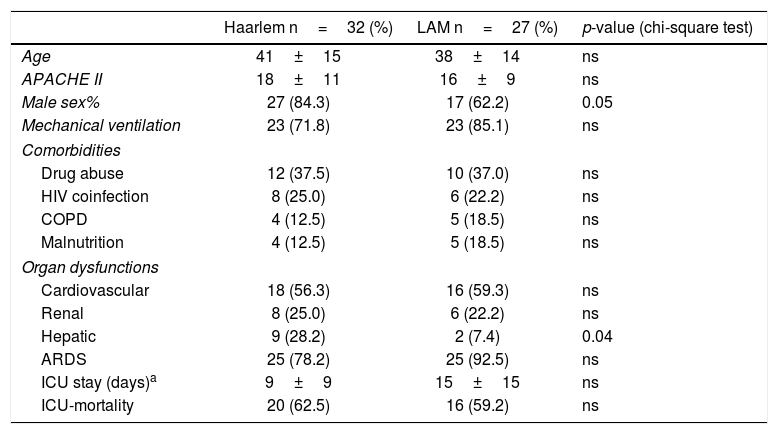
A closer look into critically ill patients is shown in Table 2, comparing demographic data and some comorbidities such as drug abuse, HIV, COPD, and malnutrition, between the main represented strains (Haarlem and LAM families). We did not find any significant differences among comorbidities between the genotype groups.
Demographic data, comorbidities, and overall outcomes comparing the Haarlem and LAM families in ICU patients with moderate to high severity score.
| Haarlem n=32 (%) | LAM n=27 (%) | p-value (chi-square test) | |
|---|---|---|---|
| Age | 41±15 | 38±14 | ns |
| APACHE II | 18±11 | 16±9 | ns |
| Male sex% | 27 (84.3) | 17 (62.2) | 0.05 |
| Mechanical ventilation | 23 (71.8) | 23 (85.1) | ns |
| Comorbidities | |||
| Drug abuse | 12 (37.5) | 10 (37.0) | ns |
| HIV coinfection | 8 (25.0) | 6 (22.2) | ns |
| COPD | 4 (12.5) | 5 (18.5) | ns |
| Malnutrition | 4 (12.5) | 5 (18.5) | ns |
| Organ dysfunctions | |||
| Cardiovascular | 18 (56.3) | 16 (59.3) | ns |
| Renal | 8 (25.0) | 6 (22.2) | ns |
| Hepatic | 9 (28.2) | 2 (7.4) | 0.04 |
| ARDS | 25 (78.2) | 25 (92.5) | ns |
| ICU stay (days)a | 9±9 | 15±15 | ns |
| ICU-mortality | 20 (62.5) | 16 (59.2) | ns |
The values represent the percentage or mean±SD.
COPD=chronic obstructive pulmonary disease, ARDS=acute respiratory distress syndrome. ns: non significant.
a Considering only patients who survived.
Comparing organ dysfunctions observed among ICU patients (with a low to high severity score) between the different genotypes, we found that patients with the Haarlem genotypes showed hepatic dysfunction more frequently than patients with LAM (p=0.04) (Table 2). The mortality observed in these patients was not statistically related to the M. tuberculosis genotype (Table 2), and no significant association between the outcome of patients with different lineages was found. As far as we know, this is the first communication relating clinical and molecular epidemiology in critically ill patients with severe TB. Furthermore, we were able to compare the main TB lineages that circulate both in the community and critical care setting, studying the association between these genotypes with clinical outcomes.
The principal finding showed that lineages belong to the Haarlem and LAM families in both studied populations, but observing a statistically significant difference between the proportions of each family among these two populations. In addition, we were able to perform a lineage comparison considering the clinical outcome in community patients.
Although, the genotype information obtained with MIRU and spoligotyping is partial (and WGS analysis could shed light about genome implications in clinical outcome), the data presented in this work suggest a more significant contribution of host factors related to the severity of the disease.
In terms of ICU mortality, neither the LAM nor the Haarlem families showed higher virulence. Factors contributing to death in the present study were more likely related to host comorbidities (in particular, drug abuse, poor nutritional status, and lack of access to health care facilities).
Although the presented series in our study is one of the largest reported5,6,13, the limited number of patients enrolled warrants a more robust statistical analysis. The study of cases should continue to add more information leading to improve the understanding of the correlation between TB lineages and virulence.
The results presented in this work represent a starting point, an in-depth study based on genomes could contribute to better understand the virulence observed in these strains and could help to improve the prognosis of these critically ill patients.
Financial supportThis work was supported by grant FSS_2_2011_1_6850 (Fondo Sectorial de Salud, Agencia Nacional de Investigación e Innovación [ANII)]. FOCEM - Fondo para la Convergencia Estructural del Mercosur (COF 03/11).
G.G., N.N., J.H. and C.R. are researchers from the Sistema Nacional de Investigadores (ANII), Montevideo, Uruguay.
Conflict of interestThe authors declare that they have no conflicts of interest.