Bovine pestiviruses are the causative agents of bovine viral diarrhea, a disease that causes severe economic losses in cattle. The aim of this study was to improve their diagnosis by developing a RT-qPCR to detect bovine pestiviruses A, B and H; and to set up a protocol for collecting, shipping and preserving bovine pestiviral RNA on filter papers. The developed RT-qPCR showed high sensitivity in detecting these viruses in different matrices: viral stocks, semen and serum samples. With regard to the possibility of using the technique to test serum pools, it was possible to identify a positive serum sample within a pool containing 30 sera. In addition to evaluating the qPCR from fresh samples, the use of filter papers to sow bovine samples was analyzed. The sampling method on two different filter papers using bovine blood drops was a useful alternative for diagnostic purposes and allowed to preserve pestiviral RNA for up to 12 months under refrigeration.
Los Pestivirus bovinos son los agentes causales de la diarrea viral bovina, una enfermedad que genera importantes pérdidas económicas en el ganado vacuno. El objetivo de este trabajo fue mejorar su diagnóstico mediante el desarrollo de una RT-qPCR para detectar los Pestivirus bovinos A, B y H y diseñar un protocolo de recolección, envío y conservación de ARN viral en papeles de filtro. La RT-qPCR desarrollada demostró alta sensibilidad en la detección de estos virus en diferentes matrices: stock viral, suero y semen. Respecto de la posibilidad de usar la técnica para testear pools de suero, fue posible identificar un suero positivo dentro de un pool compuesto por 30 sueros. Además de evaluar la qPCR en muestras frescas, se analizó el uso de papeles de filtro para sembrar muestras de bovinos. La metodología de toma de muestras en dos tipos de papeles de filtro usando gotas de sangre fue una alternativa útil para el diagnóstico y permitió conservar ARN viral por hasta 12 meses a temperaturas de refrigeración.
Bovine pestiviruses cause severe economic losses in cattle worldwide. Pestiviruses belong to the family Flaviviridae, and are single-stranded RNA viruses, classified into the genus Pestivirus, also including Pestivirus C (Classical swine fever virus) and Pestivirus D (Border disease virus)15. Two species of bovine pestiviruses (also known as Bovine viral diarrhea virus) are known: Pestivirus A and Pestivirus B (previously known as BVDV-1 and BVDV-2, respectively)29. Two decades ago, an emerging bovine pestivirus was reported28, which was recently classified as Pestivirus H (HoBi-like pestivirus)29. Each genotype is further classified into cytopathic (CP) and non-cytopathic (NCP) biotypes, according to their ability to cause damage in cell cultures11.
Bovine pestiviruses can be transmitted to susceptible animals through all the secretions of an infected animal. Persistently infected animals (PI) play a fundamental role in the transmission of the virus since they are the main source of infection through their fluids to their cohabitants. They acquire the viral infection through a NCP strain that crosses the placenta during pregnancy. From days 45 to 125 of pregnancy the immune system of the fetus is not developed; therefore, it is incapable to elicit a proper immune response. Infection in this period of time leads to the development of a PI and seronegative calf35. Thus, PI animals spread the virus through their secretions and excretions lifelong.
Pestiviruses A, B and H are the causative agents of bovine viral diarrhea (BVD), which includes a wide range of syndromes, such as diarrhea, respiratory disease, reproductive failures, embryonic mortality, abortions, malformations and hemorrhagic syndromes, depending on the age of the host and the viral strain. In addition, in PI animals, bovine pestiviruses can cause a severe syndrome called mucosal disease when they are re-infected with a CP strain of the virus12.
Identification and elimination of PI animals is essential for the control of BVD in infected herds. Furthermore, application of biosecurity measures, such as testing all animals arriving at the herd, testing semen, avoiding the contact with neighboring herds, controlling other species that could host the virus, among others, is necessary to prevent the reintroduction of pestiviruses in the herd36. In some European countries, eradication of bovine pestiviruses was accomplished by applying the mentioned biosecurity measures, including or not vaccination programs17.
Detection of PI animals requires the use of direct diagnostic techniques. Among the available assays, viral isolation is the gold standard test; however, it is expensive and takes several days to yield results, hence, being currently disused in most diagnostic laboratories. Other techniques, such as enzyme-linked immunosorbent assays, direct immunofluorescence and immunohistochemistry, can be used to detect viral antigens. As an alternative, reverse transcription-polymerase chain reaction (RT-PCR) and quantitative RT-PCR (RT-qPCR) detect viral RNA with high sensitivity31,32. qPCR has several advantages, such as fastness, and the possibility to pool samples from different animals7. Once the pools have been checked by RT-qPCR, if one of them is positive, it has to be split, and each sample must be tested individually. Compared with individual testing, the chance to test several animals as a pool is a great advantage in herds with low BVD prevalence, taking into account the saving of reagents and time consumed by technicians. Several strategies to pool samples according to the prevalence of BVD in herds have been proposed19. Currently, commercial in vitro diagnostic RT-qPCR kits are available to assure the detection of a bovine pestivirus positive animal within a pool of 50 samples; however, they may be inaccessible in some low-income countries because of their price and import constraints.
One of the critical points for molecular diagnostics is correct sample preservation. As bovine pestiviruses are very labile RNA viruses15, a cold chain must be ensured to avoid erroneous results in the RT-qPCR assay; therefore, samples must be taken and sent chilled to the laboratory as soon as possible. This is an inconvenient in herds located in places far from the diagnostic laboratories. Blood samples should be maintained at 4°C while semen should be sent in liquid nitrogen, which represents high costs and shipping logistics. A useful alternative to improve collecting and shipping samples is filter paper (cotton-based, cellulose paper), such as the Flinders Technology Associates cards (FTA® cards), especially designed to maintain nucleic acids stable at different temperatures and to protect them from UV rays25. Since microorganisms present in samples spotted onto filter papers are immediately inactivated, the FTA system is completely safe and the cards could be shipped by postal mail, at room temperature (RT). FTA cards are widely used in different areas, such as biobanking, forensic genetics and molecular epidemiology research, since they preserve nucleic acids for long time periods. They have been effective for the detection of RNA viruses (Foot-and-mouth disease virus, avian influenza virus, Porcine reproductive and respiratory syndrome virus, Rotavirus and some arboviruses)2,9,10,14,33. This technology is easy to use and non-invasive5, as a small volume of sample is spotted on a filter paper and venipuncture is not needed.
The aims of this study were to develop and validate a Sybr Green-based RT-qPCR test to assess detection and quantification of all bovine pestiviruses in serum and semen samples and to set up a strategy for collecting, shipping and preserving viral RNA on filter papers for BVD diagnosis and biobanking as a cost-effective method.
Materials and methodsVirus, plasmid and clinical samplesViral strainsA CP Pestivirus B (isolate vs253) strain cultivated in Madin Darby bovine kidney cells (MDBK) available in the Virology Institute and Innovative Technologies (IVIT) from the Research Center of Veterinary Sciences (CICVyA), INTA Castelar, was used as viral stock to standardize the RT-qPCR and to spike specimens in order to mimic clinical samples.
Semen and serum samplesTo detect RNA from bovine sera, blood samples without anticoagulant from healthy animals (pestivirus-negative by an in-house validated RT-PCR) were collected and sent to the laboratory at 4°C. Sera was obtained after a five minute-centrifugation at 1500×g and then dispensed in sterile 1.5ml tubes. The sera tubes were stored at 4°C or at −70°C in case the sample was not immediately processed.
To assemble pools, aliquots of the same volume of each bovine serum were mixed in a 1.5ml tube. The volume of each serum to be used in a pool was calculated according to the size of the pool, with a final volume of 200μl.
To detect viral RNA from semen, 250μl straws of extended semen from healthy animals (pestivirus-negative by an in-house validated RT-PCR) were shipped in liquid nitrogen and stored at −70°C. All samples were thawed at RT before processing.
Sample preparationThe amount of virus used to spike sera and semen straws was calculated based on expected PI animal viral titers (104TCID50/ml) as determined by several authors3,16. A titrated Pestivirus B stock was used to spike the samples.
Plasmid constructionFor the qPCR standardization, a plasmid containing the pestiviral 5′untranslated region (5′UTR) of the viral genome was constructed. Partial 5′UTR of a local Pestivirus A isolate (25366) (obtained by RT-PCR) was inserted into a pGemT-easy vector (Promega, USA) following the manufacturer's recommendations. Competent Escherichia coli DH5-α strains were used for transformation with the ligation product. Transformation was carried out by thermal shock, following molecular biology standard methods26. Plasmid was purified using the Wizard Plus SV Minipreps kit (Promega, USA) following the manufacturer's instructions. Plasmid identity was confirmed using restriction endonuclease digestion. Finally, the purification product was quantified by Nanodrop 2000 (Thermo Scientific, USA) and the number of copies was calculated.
Clinical samplesTwenty-eight (28) clinical samples collected from naturally infected animals that were confirmed positive by viral isolation in cell culture and subsequently direct immunofluorescence, were used to ensure viral RNA detection by the assay. The specimens consisted of 4 intestines, 7 fetuses, 3 nasal swabs, one ocular swab, 2 esophagus, one lung, 2 spleens and 8 blood samples (provided and diagnosed by Laboratorio Azul and Specialized Veterinary Diagnostic Service at INTA Balcarce, Argentina).
qPCR standardizationqPCR standardization was performed according to the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines4.
RNA extractionRNA extraction was performed using the High Pure PCR Template Preparation kit (Roche, Switzerland) following the manufacturer's instructions. Positive and negative controls were included, consisting of 200μl of Pestivirus B (isolate vs253) and 200μl of elution buffer, respectively. The obtained RNA was immediately stored at −70°C until processing.
Retro-transcription and qPCR reactionRT-qPCR reactions were carried out simultaneously using the iTaq Universal SYBR Green One-Step Kit (Bio-Rad, USA). A 10μl final volume mix was prepared with 5μl of iTaq Universal SYBR Green reaction mix, 0.125μl of iScript reverse transcriptase, 0.25μl of forward and reverse primer (Genbiotech, Argentina), 2.625μl of nuclease free water (Biodynamics, Argentina) and 2μl of RNA obtained in a previous step. The cycling program consisted of 10min at 50°C for reverse transcription reaction, 1min at 95°C for polymerase activation and DNA denaturation, followed by 40 cycles of 15s at 95°C and 1min at 60°C, finalizing with a 0.5°C increment from 65 to 95°C every 2s, using StepOne equipment (Applied Biosystems, USA). In each assay, positive and negative controls were included.
Primer selection and concentration adjustmentPrimer pairs 189–389 were used to develop the qPCR18. Primer sequences were 5′-AGTCGTCAR(A/G)TGGTTCGAC-3′ and 5′-TCCATGTGCCATGTACA-3′ for 189 (forward primer) and 389 (reverse primer), respectively and amplify a 201bp sequence of the 5′UTR of bovine pestiviral genome. The expected amplicon was verified using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and aligned with reference NADL strain (GenBank accession number M31182) between positions 190 and 390 of the complete sequence.
To establish the most efficient dilution of primers to set up the test, 10 folded dilutions of the plasmid were prepared and several concentrations of primers were assessed (390nM, 420nM, 450nM, 470nM, 525nM, 535nM and 540nM). Primer efficiency was confirmed for each experiment using the formula: E=−1+10˄(−1/slope)22, whereby a slope close to −3.32 indicates 100% efficiency. This ensured that PCR products were amplified at an efficient rate and experiments were comparable among them.
Analytical sensitivity and linear dynamic range of detectionLimit of detection (LOD) was calculated for the different matrices: plasmid, viral stock and spiked serum and semen. For that purpose: 10-fold dilutions were performed, from 109 to 100 copies of the plasmid (3 replicates) and from 105 to 100 copies of the viral stock, sera and semen (12 replicates). In addition, 3 replicates of pools of bovine sera containing 10, 20, 30, 40 and 50 individual samples (one serum of each pool was spiked to mimic a PI sample) were assessed in order to determine the efficiency in pooling sera. The linear range of the qPCR was established by a standard curve with 10-fold dilutions of the plasmid from 2.5 to 2500000000 copies.
Assay precisionReplication experiments were carried out to test positive controls (plasmid and viral stock) and to compare cycle threshold (CT) results in order to ensure precise detection of the target sequence. CT values of 25 repetitions of viral stock and 10 repetitions of 5′UTR plasmid (processed in triplicate) were registered. Mean, standard deviation, weighted average variation coefficient (CVpp) and maximum random error allowed (CVa%) were calculated.
Analytical specificityBovine respiratory syncytial virus (BRSV), Bovine parainfluenza-3 (PI-3), Bovine herpesvirus-5 (BHV-5), Bovine herpesvirus-1 (BHV-1), Blue tongue virus (BTV), Bovine Rotavirus type A (RVA) and Mycoplasma spp. were used to evaluate the specificity of the test. In addition, several bovine pestiviral strains available at the IVIT, INTA Castelar, were used to check the ability of the qPCR to identify all possible viral variants that were detected in Argentina. Viral isolates and reference strains included: 13 strains of Pestiviruses A (4 from sub-genotype a and 9 from sub-genotype b), 4 strains of Pestivirus B and 5 strains of Pestivirus H.
Inter-laboratory assaysDNA aliquots of plasmid and negative controls, as well as reagents and instructions to run the test were sent to three different laboratories to perform the qPCR. Two of them were located in the IVIT Institute and the other one was from a private veterinary diagnostic laboratory. Involved laboratories have trained technicians and separate areas to prepare PCR mixes and sow samples. The assay was also carried out in equipment available in these laboratories: iQ5 (Bio-Rad, USA), ABI7500 (Applied Biosystems, USA) and MyGo (IT-IS Life Science, Ireland) thermal cyclers.
Data analysisData analysis was carried out using StepOne Software version 2.3.
Filter paper sample processingStorage of filter papers spotted with spiked bloodTwo types of filter papers were used throughout the experiments: FTA cards (Whatman, UK) and cellulose 3mm chromatography paper (GE Healthcare, USA). Since FTA cards are designed for nucleic acid conservation over long time periods, they were used for conservation purposes. Conversely, chromatography paper, commonly used for other purposes and less expensive than the former, was evaluated for rapid laboratory tests in cases of suspected BVD in the field. This means, preserving RNA for shorter periods of time, allowing the collection and shipping of samples to the laboratory at room temperature.
Whole-blood samples were collected from healthy bovines, and then spiked with a titrated Pestivirus B viral stock as previously described so that there were 50000 copies of RNA in a 50μl drop, according to PI animal viral titers.
Five repetitions of 50μl of spiked blood drops were spotted on chromatography paper using micropipettes and sterile tips. Then, the drops were dried at RT for 4h and finally stored at RT (15–25°C), 4°C, −20°C and −70°C for one and 7 days and 6, 12 and 18 months. The same methodology was performed on FTA cards: 5 drops of spiked blood were spotted for each temperature and stored for 12 and 18 months. The spiked filter papers were stored at different temperatures in zipped bags with desiccant to prevent humidity damage.
Once storage time expired, RNA was eluted. Briefly, four punches in each blood drop were cut with a 6mm puncher and placed in 1.5ml tubes containing 220μl of minimum essential medium (MEM). The tubes were then kept in a shaker for 30min at RT and aliquots of 200μl were taken to proceed with RNA extraction and RT-qPCR described above including viral stock used as a positive control.
In order to prevent cross-contamination among samples and replicates, a protocol recommended in another study was followed. Briefly, the puncher was decontaminated between samples by dipping it in isopropyl alcohol for a minute. Later, the puncher was dried with absorbent paper and five punches were cut in a clean, new chromatography paper27.
Limit of detection in filter papersTo assess the LOD in blood spotted on filter papers, samples were spiked with viral stock at several concentrations and spotted on filter papers. For this purpose, two replicates of 50μl drops of blood containing 100, 1000, 5000, 10000, 20000, 40000 and 80000 viral particles were spotted on chromatography paper and later processed as mentioned above.
Recovering RNA from clinical samplesBlood drops of two PI animals were spotted on chromatography paper, dried up at RT for 4h and then stored at 4°C for 1 day and 6, 12 and 18 months. Once storage time expired, samples were processed as previously mentioned.
Statistical analysisAiming to compare the efficiency in recovering RNA between both types of papers at 12-month storage, a statistic analysis was conducted using the T-student and Fisher's tests. CT values and positive/negative results for each repetition stored at 4°C and −20°C were used for the T-Student and Fisher's tests, respectively.
Internal control to monitor nucleic acid extraction and PCR inhibitorsThe efficiency of nucleic acid extraction and the presence of inhibitors for PCR amplification in semen and blood spotted on filter papers, fresh semen and tissue samples was checked by a qPCR assay using a set of primers that specifically amplify a fragment of the constitutive bovine GAPDH gene. For this purpose, GAPDH forward and reverse primer pairs24 were used.
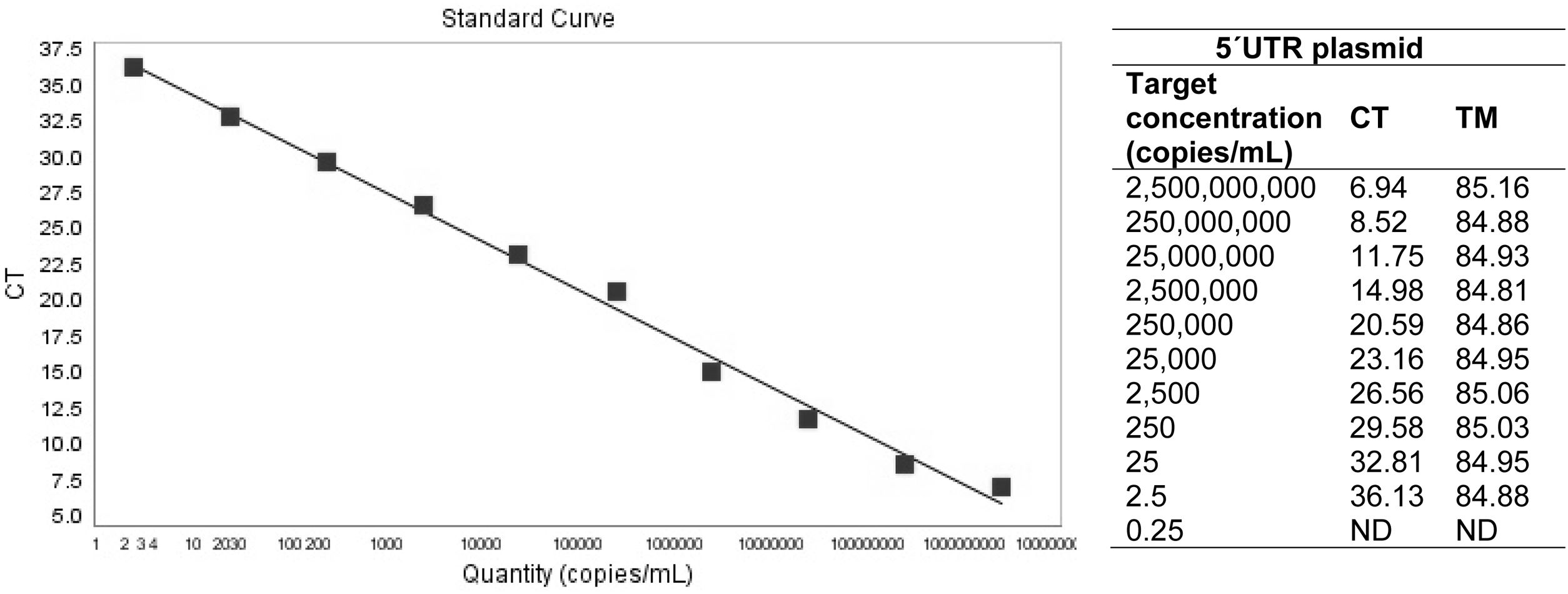
ResultsRT-qPCR standardizationWith regard to primer concentration adjustment, the best efficiency was observed in 470nM (0.47μM). The linear range of the PCR was from 2.5×109 to 2.5×101 DNA copies/ml, with an efficiency of 97.3%, a slope of −3.389 and regression coefficient of 0.994. Based on the calculations made on the plasmid, the test detected 2.5 copies (LOD) (Fig. 1). Regarding the qPCR sensitivity for viral stocks, the qPCR amplified up to five copies of viral RNA. Among the sera and semen matrices, the test was able to detect up to 50 RNA copies (Table 1a). Furthermore, the RT-qPCR test was able to detect a positive serum in a pool containing 30 bovine sera (Table 1b).
(a) Number of copies detected by the qPCR in different matrices: virus stock (Pestivirus B vs253), serum and semen straws. (b) CT (cycle threshold) mean for three replicates of the different pool sizes evaluated.
| (a) Target concentration (copies/ml) | Viral stock | Serum | Semen straws |
|---|---|---|---|
| 500000 | + | + | + |
| 50000 | + | + | + |
| 5000 | + | + | + |
| 500 | + | + | + |
| 50 | + | + | + |
| 5 | + | ND | ND |
| (b) Pool size (number of individual sera within the pool) | CT |
|---|---|
| 10 | 30.3 |
| 20 | 30.7 |
| 30 | 32.2 |
| 40 | ND |
| 50 | ND |
ND: not detected.
Results are expressed as 95% agreement between 12 replicates.
In regard to specificity, the qPCR did not amplify unrelated microorganisms (BRSV, PI-3, BHV-1, BHV-5, BTV, RVA and Mycoplasma spp.). Furthermore, the test was able to detect all bovine pestivirus strains evaluated. Melting temperature (TM) values for each bovine pestiviral strain assayed is shown in Table 2, the specific range of TM went from 84.69 to 85.78°C. The precision analysis of the 5′UTR plasmid and viral stocks showed a relatively low standard deviation and poor dispersion of CT results with respect to the mean (Table 3). Concerning the inter-laboratory assays, it was possible to detect the plasmid in the three different laboratories using several cyclers, with CT values ranging from 13.92 to 22.
Melting temperature (TM) of bovine pestiviral strains assayed by the RT-qPCR.
| Strain | TM (°C) |
|---|---|
| Bovine Pestivirus Aa Singer | 85.41 |
| Bovine Pestivirus Aa MF120553 | 85.50 |
| Bovine Pestivirus Aa 88625 | 85.78 |
| Bovine Pestivirus Aa 71267 | 85.2 |
| Bovine Pestivirus Ab 63588 | 85.08 |
| Bovine Pestivirus Ab 90611 | 85.59 |
| Bovine Pestivirus Ab MF120552 | 85.10 |
| Bovine Pestivirus Ab 59473 | 85.32 |
| Bovine Pestivirus Ab 83532 | 85.62 |
| Bovine Pestivirus Ab 73611 | 84.81 |
| Bovine Pestivirus Ab MF120594 | 85.22 |
| Bovine Pestivirus Ab 78256 | 85.42 |
| Bovine Pestivirus Ab 25366 | 85.32 |
| Bovine Pestivirus B 76/08 | 85.36 |
| Bovine Pestivirus B vs253 | 85.59 |
| Bovine Pestivirus B MF120585 | 85.44 |
| Bovine Pestivirus B MF120586 | 85.61 |
| Bovine Pestivirus H LV210185K/13 | 84.69 |
| Bovine Pestivirus H MK017821 | 84.80 |
| Bovine Pestivirus H MH992643 | 85.58 |
| Bovine Pestivirus H #48 | 85.1 |
| Bovine Pestivirus H #51 | 84.92 |
Mean, standard deviation, repetitions number (n), weighted average variation coefficient (CVpp) and maximum random error allowed (CVa%) of positive controls repetitions.
| Mean | Standard deviation | n | CVpp | CVa% | |
|---|---|---|---|---|---|
| 5′UTR plasmid (2.5×103 copies) | 21.36 | 1.49 | 10 | 14.94 | 8.96 |
| Pestivirus B vs253 (5.62×106 copies) | 13.78 | 1.94 | 25 | 10.63 | 6.38 |
All clinical specimens collected from naturally infected animals which were positive to conventional gold standard methods (viral isolation followed by direct immunofluorescence) were positive when analyzed through the developed RT-qPCR, with CT values ranging from 15 to 35 (Supplementary material 1).
Finally, the GAPDH gene, which was used to monitor nucleic acid extraction and possible presence of PCR inhibitors in blood and semen spotted on filter papers, fresh semen and tissue samples, were positive in all runs, with TM results ranging from 80 to 81°C and homogeneous CT values (20±3) for all specimens.
Pestiviral detection on filter papersFilter papers spotted with spiked bloodAs expected, when the storage period increased, the RNA recovering percentage among repetitions decreased. For instance, one and 7-day storage of blood drops on chromatography papers yielded a higher recovery percentage with respect to positive control CT when compared with 6 and 12-month storage. Regarding storage temperatures, in general, 4°C, −20°C and −70°C were more effective in preserving RNA that RT, with a more marked trend as storage time increased (Table 4a).
Replicates cycle threshold (CT) means, number of positive determinations and recovery percentage at different time-points and temperatures for spiked blood spotted on filter papers. (a) From chromatography paper. (b) From FTA cards.
| RT | 4°C | −20°C | −70°C | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CT mean | PD | SD | CT mean | PD | SD | CT mean | PD | SD | CT mean | PD | SD | |
| (a) | ||||||||||||
| 1 day | 27.14 | 5 | 2.09 | 25.58 | 4 | 0.57 | 25.64 | 5 | 1.98 | 27.23 | 4 | 1.78 |
| Recovery % | 84.16 | 89.29 | 89.08 | 83.88 | ||||||||
| 7 days | 28.77 | 5 | 4.67 | 28.67 | 5 | 0.99 | 28.47 | 5 | 1.12 | 26.26 | 5 | 2.12 |
| Recovery % | 79.39 | 79.67 | 80.22 | 86.98 | ||||||||
| 6 months | 32.1 | 3 | 1.18 | 25.12 | 3 | 1.73 | 24.83 | 5 | 0.84 | 22.02 | 5 | 0.99 |
| Recovery % | 48.35 | 61.78 | 62.51 | 57.67 | ||||||||
| 12 months | ND | 0 | – | 27 | 4 | 0.6 | 26.87 | 4 | 0.96 | NP | ||
| Recovery % | 0 | 67.81 | 68.14 | |||||||||
| (b) | ||||||||||||
| 12 months | ND | 0 | – | 27.11 | 3 | 0.55 | 28.52 | 3 | 8.78 | NP | ||
| Recovery % | 0 | 67.81 | 64.20 | |||||||||
ND: not detected; NP: not processed.
When analyzing the samples stored for 12 months, results were similar in both types of papers (chromatography paper and FTA cards) at 4°C and −20°C storage temperatures (Table 4b). No significative differences were identified between both types of papers at 12-month storage at 4°C (p=0.865) and at −20°C (p=0.776) using the T-Student test, neither at 4°C (p=0.500) nor at −20°C (p=0.500) using the Fisher's test.
Unfortunately, it was not possible to process the samples stored at −70°C. At 18 months, results were negative for all storage temperature variables in both types of papers.
Limit of detection in chromatography paperWith regard to the LOD in blood samples, the qPCR was able to detect up to 1000 copies of viral RNA (Table 5).
Recovering RNA in papers from clinical samplesBlood drops from two PI animals spotted on chromatography paper and stored for one day and 6 and 12 months at 4°C yielded positive results by the developed RT-qPCR, suggesting that after one year of storage at 4°C, there was no significant degradation of the viral RNA on the chromatography paper for both PI animals. CT values for the samples for one day storage were 28.39 and 32.9; for 6-month storage were 23.93 and 33.75; finally, for 12-month storage were 29.93 and 27.56, respectively. On the contrary, results were negative at 18-month storage time.
DiscussionThe developed RT-qPCR was able to detect the viral RNA in all bovine pestiviral strains tested and showed high specificity. A major advantage is the ability to detect Pestivirus H strains, which can be useful in areas where Pestivirus H are co-circulating with Pestivirus A and B. Furthermore, complete agreement was observed between RT-qPCR and viral isolation results of all clinical samples tested. In pooled sera, the test was able to detect a positive animal among 30 sera, which is a significant advantage when carrying out BVD control in low-prevalence herds since it allows to reduce the number of assays. However, in herds where BVDV prevalence is high, there is an increased chance that any single pool will test positive if the pool size is too large, requiring additional testing to identify the viremic individuals19. Thus, prevalence estimation should be considered when laboratories use the pooling-strategy, especially in regions with high BVD prevalence30. Moreover, when the pool contains a large number of individual samples, the dilution effect can cause false negatives. This research has provided an in-house validated RT-qPCR that can replace commercially available kits, which represent a great trading cost for some countries, and can be useful for veterinary diagnostic laboratories equipped with the infrastructure to perform qPCR assays.
Because of the SYBR Green-based detection system, the developed RT-qPCR presents some advantages: it is sensitive, easy to use and inexpensive. However, it is important to recognize the presence of any double-stranded DNA (e.g., primer-dimers) that can cause false positive results. This inconvenient can be sorted including an analysis after the amplification run, using the melting temperature of the expected amplicon, which allows the discrimination of the target amplicon to undesired products that can interfere with the results23. In addition, using One-Step for retro-transcription and PCR reaction simultaneously reduces the costs, time consumed and possibilities of contamination of the assay.
Diagnostic laboratories routinely use blood or serum for BVD analyses. In terms of practicability in the use of filter papers at the point of care, whole blood is more suitable than serum since it avoids the centrifugation step. Although semen is not used as a sample of choice to diagnose bovine pestiviruses, it is useful for the control of BVD in breeding herds20,21; however, our group tested the same methodology using semen straw samples spotted on filter papers without achieving satisfactory results (Supplementary material 2).
Concerning the collection and conservation of bovine pestiviral RNA on filter paper, the literature is scarce. One article describes the use of several filter papers to preserve RNA up to six months in blood and sera samples from PI animals at RT, 4°C and −18°C34. In this study, the authors evaluated four different types of papers: classical filter papers, Whatman paper No. 1, nitrocellulose membrane and HYBOND™-M nylon membrane. Results were similar for all papers tested except for classical filter papers, which yielded lower PCR products. However, the authors used an end-point RT-PCR assay and RNA isolation was carried out directly from the filter papers, avoiding the elution step. Unfortunately, that strategy did not yield good results in our experiments (data not shown). Another study was reported in FTA cards for the detection of several viral agents involved in the bovine respiratory complex, including bovine pestivirus, where viral RNA from respiratory tract swabs was stored for 14 days at temperatures between −27 and 46°C. The article compared the RNA recovery from specimens in viral transport medium and FTA fixed samples, which proved 100% agreement13. Unlike the former, in this paper, the authors used a qPCR assay and carried out an elution protocol before RNA extraction. Finally, a third article describes the use of FTA cards to spot nasal swabs and blood samples from PI animals that were immediately processed by real time-PCR without storage. In this paper, the authors compared the mentioned sampling method to fresh samples, and observed good agreement between both, except for an increase in CT values in FTA samples compared with fresh samples, which was not considered significant given the high viral load in PI samples8.
Our study suggests that chromatography papers are a useful alternative for collecting and shipping blood samples for BDV diagnostic purposes, as a cost-effective method. However, for longer storage time (6 and 12 months), the recovery percentages decreased, although RNA was still detected, meaning that the biobanking purposes are not suitable for more than 12-month storage at any temperature with the protocols evaluated in this experiment. Nevertheless, the fact that pestiviral RNA was amplified in PI animal blood stored up to 12 months in chromatography paper, suggests that, although the recovery percentage was lower as the storage time progressed, it was still possible to recover RNA in blood samples from PI animals kept at refrigeration temperature.
FTA cards contain chelating agents and a free-radical trap designed to deal with atmospheric pollutants, thus protecting the entrapped nucleic acids for at least six years at room temperature1. However, in our work, chromatography paper -which lacks these components-, yielded similar results. Moreover, for 12-month storage, drops spotted on chromatography paper showed slightly better results than on FTA cards, considering the number of positive repetitions.
When collecting samples, we recommend drying the drop spotted on filter papers at room temperature for at least 4h (depending on ambient humidity), and sending them to the laboratory in individual zipped bags with a desiccant, such as silica gel. For shipping, refrigeration is not necessary.
The possibility of storing viral nucleic acids on filter papers constitutes a breakthrough in veterinary practice. The chance to collect samples directly on filter papers from herds where the disease is suspected could be very helpful, since in this kind of matrix, samples do not require immediate shipping or refrigeration and more importantly, they do not need to be processed instantly6. This methodology is mostly advantageous when performing molecular studies, since it guarantees nucleic acid stability at a wide range of temperatures and filter papers can be used with a variety of samples. Compared with the traditional method for collecting, shipping and storing samples, filter paper methodology represents a major improvement especially in large countries with poor infrastructure for shipping samples and also for those lacking laboratories which run this type of assays in every state or province.
An advantage of using chromatography paper as a method for collecting samples is its cost. Every sheet of paper can hold approximately 25 drops of sample, with a value of USD 66.55 per 100 sheets. In contrast, only four drops of sample can be spotted on every FTA card, with an approximate cost of USD 308 (depending on the provider) per 25 sheets.
Our study demonstrated that an economical filter paper, such as chromatography paper, is an appropriate alternative for collecting, shipping and storing blood samples for BVD diagnosis, being the results yielded as good as those using FTA specialized cards. The aforementioned is valid, as long as it is accompanied by a highly sensitive technique such as the RT-qPCR reported in this study.
Further studies using this methodology applied to other viruses are warranted, especially for diseases relevant to bovine health, which require mandatory control and are economically important for local production in order to improve sampling, shipping and laboratory processing conditions.
FundingFinancial support for the execution of this study was provided by Fundación ArgenINTA.
Conflict of interestThe authors declared no conflict of interest with respect to the research, authorship, and/or publication of this article.
The technical assistance of Maximiliano Jordan, Emiliano Nicodemo and Darío Malacari is greatly appreciated.