To conduct a study of Staphylococcus aureus carriage in members of a livestock-farmer's family with different degrees of animal contact, and to characterize the recovered isolates.
MethodsNasal samples from 11 members of the family were taken in three sampling periods (every six months) (n=31), and 9 skin samples from superficial lesions were also obtained in 5 of them. Samples were analyzed for S. aureus susceptible (MSSA) and resistant to methicillin (MRSA). S. aureus isolates were tested for antibiotic-resistance phenotype and genotype and for the detection of virulence and IEC-system genes. Molecular typing of isolates was also performed (spa- and multilocus-sequence typing).
ResultsEighteen S. aureus isolates were recovered (1 MRSA and 17 MSSA) in the 40 samples analyzed. S. aureus was detected in nasal and skin samples of 7/11 and 4/5 of tested humans, respectively. The MRSA strain was detected in the skin lesion of a farmer with high animal contact, and carried the mecC gene, and was typed as ST130-CC130-t843. The 17 MSSA isolates were ascribed to 9 different spa-types and sequence types included in the clonal complexes CC22, CC30, CC45, CC121, and in the livestock-associated lineages CC9 and CC133. Six strains harbored eta or tsst-1 genes. Three of 18 strains lacked the immune-evasion-cluster (IEC) genes (MRSA-ST130, MSSA-ST1333, and MSSA-ST133), and the remaining isolates were ascribed to IEC type-A or -B.
ConclusionsAnimal-associated S. aureus lineages were detected in samples of the farmer's family, highlighting the detection of MSSA-CC133 and mecC-MRSA-ST130.
Estudiar la presencia de S. aureus en muestras nasales y cutáneas de los miembros de una familia de granjeros, con distinto nivel de contacto con ganado, y caracterizar los aislados obtenidos.
MétodosSe recogieron 3 muestras nasales (1 cada 6 meses) de los 11 miembros de la familia de granjeros (n=31) y 9 muestras cutáneas de pequeñas lesiones de 5 de ellos, para aislamiento de S. aureus, tanto sensible (SASM) como resistente a meticilina (SARM). Se realizó la caracterización molecular de los aislados (spa- y multi-locus-sequence-typing) y se estudiaron sus fenotipos y genotipos de resistencia, y su contenido en genes de virulencia y del clúster de evasión-inmune-humano (IEC).
ResultadosSe obtuvieron 18 aislados de S. aureus (1 SARM y 17 SASM) en las 40 muestras estudiadas. De las personas examinadas, 9/11 fueron portadoras de S. aureus: 7/11 en muestras nasales y 4/5 en cutáneas. La cepa SARM fue aislada en una lesión cutánea de un granjero, portaba el gen mecC y se tipó como ST130-CC130-t843. En las 17 cepas SASM se detectaron 9 tipos de spa y 9 secuencias-tipo, adscritas a los complejos clonales CC2, CC30, CC45, CC121 y a los asociados con ganado CC9 y CC133. Seis cepas portaron los genes eta o tsst-1. Tres de las 18 cepas carecían de los genes del sistema de evasión inmune (IEC) (SARM-ST130, SASM-ST1333 y SASM-ST133), y el resto presentaron IEC-A o -B.
ConclusiónSe detectaron líneas genéticas de S. aureus asociadas a animales en la familia de granjeros, destacando la detección de SASM-CC133 y SARM-ST130-mecC.
Staphylococcus aureus is a microorganism that can be found in the natural microbiota of humans and animals, mainly colonizing skin and nasopharynx.1 It has been considered as one of the most common opportunistic pathogens, able to cause infections of varying severity.2,3 In some situations, these infections are caused by S. aureus with methicillin-resistance determinants as mecA gene, which confers resistance to most β-lactams. Thus, some genetic lineages, as the clonal complexes CC5, CC22, CC30 or CC45, have been associated with hospital environments (HA-MRSA) with high capacity to cause nosocomial infections and to carry a large set of antibiotic-resistance genes.4 On the other hand, genetic lineages as CC8 or CC80 and CC30, among others, have been related to community environments (CA-MRSA), known for its high content of virulence factors but low levels of resistance.5 However, the differences between HA-MRSA and CA-MRSA isolates have become diffuse, and clonal lineages considered community associated are being related to hospital environments and vice versa.
Moreover, in the last years, other genetic lineages belonging to CC398, CC1, CC9, CC130 or CC133, known as livestock-associated MRSA (LA-MRSA), have emerged as an important zoonotic problem.6,7 They have been increasingly detected in animals or humans, either as colonizer or causing infections.6,8 In 2011, a new mecA homologue was described in the S. aureus isolate LGA251 from an epidemiological study of bovine mastitis in England.10 This new resistance determinant shared homology of 70% over the mecA and was named as mecALGA251,9 and renamed in 2012 as mecC10 and was integrated into the new chromosomal cassette SCCmec-XI.11
There is special interest to know the genetic lineages of S. aureus that are circulating in livestock environments in order to track its evolution, particularly those genetic lineages with high zoonotic potential, as CC398, CC130 or CC133. For this aim, the use of molecular techniques, such as spa-, agr-, or MLST typing, is very useful. A recent important tool for discriminating possible human or animal origin of isolates seems to be the determination of the human immune evasion cluster (IEC). The IEC constitutes a set of genes that enhances colonization by S. aureus allowing evasion of the first barriers of the human immune defenses, and increasing their invasive abilities.2,12
The objective of the present study was to perform a longitudinal study of S. aureus carriage in 11-member family of livestock farmers with different animal contact degree. The purpose was to carry out the molecular characterization of the recovered S. aureus isolates to determine the genetic lineages circulating and to study their resistance and virulence genes content as well as their IEC system.
Material and methodsSampling, bacterial isolation and molecular typing of isolatesA total of 11 human volunteers of a family related to a livestock farm and with different animal contact degree were included in this study: H, high contact: three farmers which worked continuously in the livestock farm; I, intermediate contact: three individuals which lived in a house in the livestock farm and worked sporadically on it; L, low contact: the remaining five individuals which cohabited with these six continuous and sporadic workers of the livestock farm. This farm was located in La Rioja region, Northern Spain, and had different types of food-animals (mainly veal calves, but also sheep, goats or horses), in addition to pet animals (dogs and cats).
Nasal samples of these 11 individuals were taken in three periods every six months (1: April 2013; 2: October 2013; 3: April 2014), and a total of 31 nasal samples were obtained (in two of the individuals only two nasal samples could be recovered). Moreover, five of the individuals presented minor skin lesions (infected eczemas or small superficial wounds) and nine samples of these origins were obtained. Individuals (age range: 26–61; 45% men and 55% women) had neither received antibiotics nor had relation with the hospital environment during the study, except for one individual who had a long-term injury in the foot, exacerbated by diabetes (case 4; Table 1). Informed consent was obtained from all individuals included in this study with adherence to ethical standards.
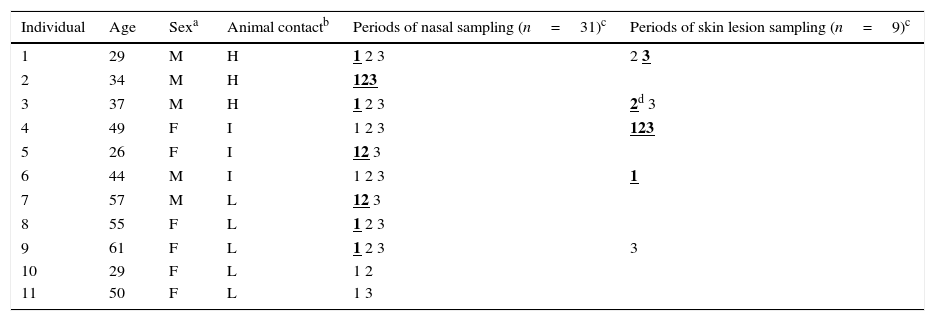
Characteristics of the 11 tested individuals and periods of sampling.
| Individual | Age | Sexa | Animal contactb | Periods of nasal sampling (n=31)c | Periods of skin lesion sampling (n=9)c |
|---|---|---|---|---|---|
| 1 | 29 | M | H | 1 2 3 | 2 3 |
| 2 | 34 | M | H | 123 | |
| 3 | 37 | M | H | 1 2 3 | 2d 3 |
| 4 | 49 | F | I | 1 2 3 | 123 |
| 5 | 26 | F | I | 12 3 | |
| 6 | 44 | M | I | 1 2 3 | 1 |
| 7 | 57 | M | L | 12 3 | |
| 8 | 55 | F | L | 1 2 3 | |
| 9 | 61 | F | L | 1 2 3 | 3 |
| 10 | 29 | F | L | 1 2 | |
| 11 | 50 | F | L | 1 3 |
Nasal and skin-lesion swabs were directly inoculated in Brain–Heart-Infusion (BHI, Becton–Dickinson) broth containing 6.5% NaCl and incubated at 37°C for 24h. Then, 100μL of this culture was seeded on Mannitol Salt Agar (MSA, Becton–Dickinson) and Oxacillin-Resistance-Screening-Agar-Base (ORSAB, OXOID) supplemented with 2mg/L of oxacillin, and the plates were incubated at 37°C for 24–36h. Presumptive S. aureus and MRSA colonies were selected in MSA and ORSAB plates, respectively. Two S. aureus isolates per positive plate were initially selected, but only isolates with different antimicrobial-resistance phenotype or different spa type per sample were further characterized. Identification of S. aureus and MRSA was performed by conventional methods (DNAse assay, catalase and Gram staining) and confirmed by specific PCRs of the nuc (S. aureus specific thermonuclease) and mecA or mecC genes.13,14S. aureus isolates were typed by: spa (www.ridom.com), agr, and multilocus sequence typing (MLST) (saureus.mlst.net).11spa- and agr-typing were performed for all S. aureus isolates. MLST was only performed in one isolate per each different spa type and SCCmec typing was carried out in methicillin resistant strains. All isolates were ascribed to a specific clonal complex (CC) (saureus.mlst.net/eburst).
Susceptibility testing and detection of resistance genesThe resistance phenotype to 16 antibiotics (penicillin, oxacillin, cefoxitin, tetracycline, erythromycin, clindamycin, ciprofloxacin, gentamicin, tobramycin, streptomycin, kanamycin, linezolid, mupirocin, fusidic acid, cloramphenicol and trimethoprim-sulfamethoxazole) was determined by the disk-diffusion method.15,16 In addition, the presence of 26 antibiotic-resistance genes [including blaZ, blaZ-SCCmec-XI, mecA, mecC, tet(K), tet(L), tet(M), msr(A), msr(B), mph(C), erm(A), erm(B), erm(C), erm(F), erm(T), lnu(A), lnu(B), ant(4′)-Ia, aph(3′)-IIIa, aac(6′)-aph(2”), aad(A), aad(E), str, fus(B), fus(C), and mup(A)] was analyzed by PCR.6,14
Detection of virulence genes and immune-evasion-cluster (IEC) genesThe virulence profile analysis was performed by PCR for detection of the following genetic determinants: lukF/lukS-PV (Panton-Valentine leukocidin), tsst-1 (toxic shock syndrome toxin), eta, etb, etd, and etd2 (exfoliative toxin A, B, D and D2 respectively). In addition, the genes encoding leukocidines DE (luk-DE) and M (lukM), the biofilm-associated protein (bap), and the collagen-binding protein (cna) were also studied by PCR.6
The five genes that comprise the IEC cluster (scn, chp, sak, and sea or sep) were studied by PCR. Depending on the presence or absence of these genes and their different combinations, S. aureus isolates were classified into 7 different IEC types according to patterns previously described.12 The scn gene is mandatory for the consideration of the IEC types.
ResultsPrevalence of S. aureus detected in this studyTable 1 shows the characteristics of the 11 members of the family studied, as well as the samples and periods analyzed. S. aureus was detected (either in nasal or skin samples) in 9 of the 11 individuals tested, 6 of them worked continuously or lived in the farm and the other three cohabited with them. S. aureus was detected in nasal and skin samples of 7/11 and 4/5 tested humans, respectively.
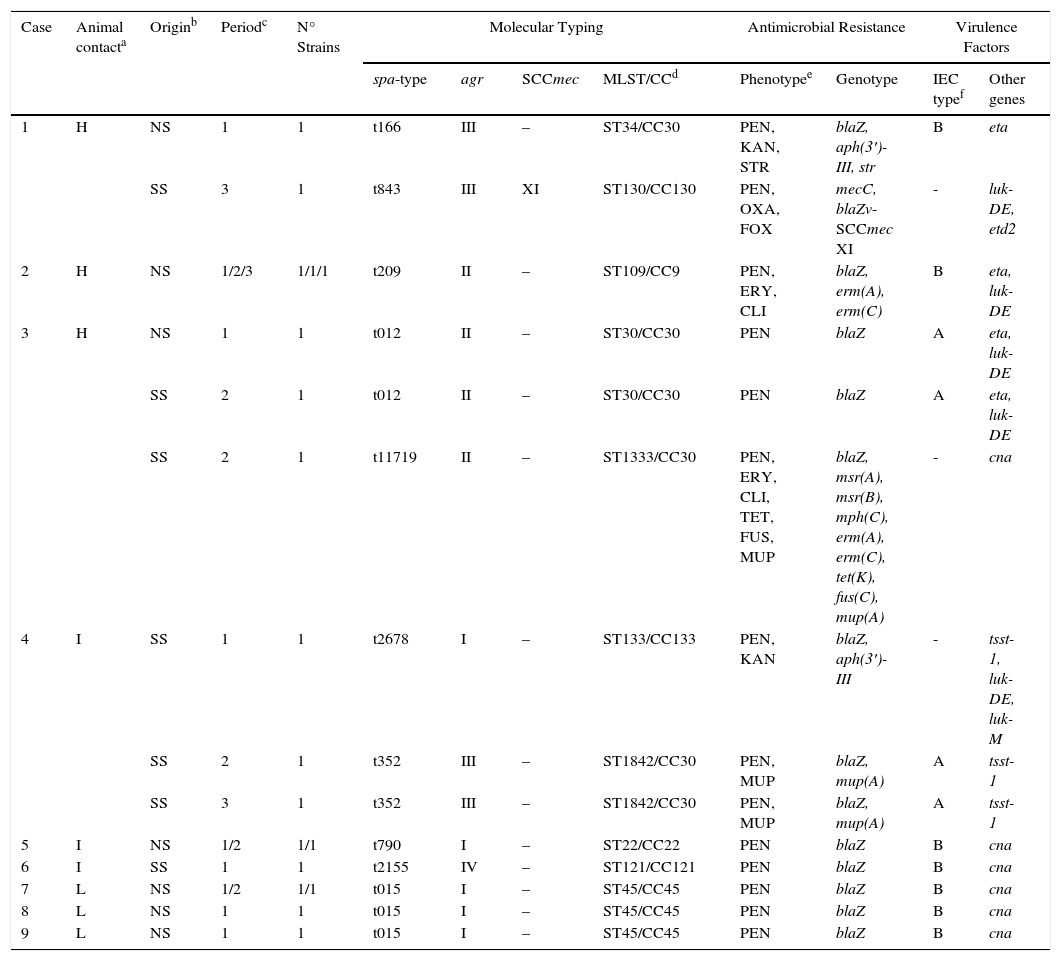
S. aureus isolates were found in 17 of the 40 samples analyzed (42.5%) that corresponded to 35.5% of nasal samples (11/31) and 66.6% of the skin samples (6/9). In one skin sample (case 3), two isolates with different resistance phenotype were identified and both were characterized. Table 2 shows the molecular characteristics of 18 studied isolates. 11 of them were of nasal origin and 7 from superficial skin lesions. Most of these isolates (17 of 18) were MSSA, but one MRSA isolate was also detected.
Characteristics of the 18 S. aureus strains recovered in this study from nasal or skin lesion samples of 9 of the 11 tested individuals.
| Case | Animal contacta | Originb | Periodc | N° Strains | Molecular Typing | Antimicrobial Resistance | Virulence Factors | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| spa-type | agr | SCCmec | MLST/CCd | Phenotypee | Genotype | IEC typef | Other genes | |||||
| 1 | H | NS | 1 | 1 | t166 | III | – | ST34/CC30 | PEN, KAN, STR | blaZ, aph(3′)-III, str | B | eta |
| SS | 3 | 1 | t843 | III | XI | ST130/CC130 | PEN, OXA, FOX | mecC, blaZv-SCCmec XI | - | luk-DE, etd2 | ||
| 2 | H | NS | 1/2/3 | 1/1/1 | t209 | II | – | ST109/CC9 | PEN, ERY, CLI | blaZ, erm(A), erm(C) | B | eta, luk-DE |
| 3 | H | NS | 1 | 1 | t012 | II | – | ST30/CC30 | PEN | blaZ | A | eta, luk-DE |
| SS | 2 | 1 | t012 | II | – | ST30/CC30 | PEN | blaZ | A | eta, luk-DE | ||
| SS | 2 | 1 | t11719 | II | – | ST1333/CC30 | PEN, ERY, CLI, TET, FUS, MUP | blaZ, msr(A), msr(B), mph(C), erm(A), erm(C), tet(K), fus(C), mup(A) | - | cna | ||
| 4 | I | SS | 1 | 1 | t2678 | I | – | ST133/CC133 | PEN, KAN | blaZ, aph(3′)-III | - | tsst-1, luk-DE, luk-M |
| SS | 2 | 1 | t352 | III | – | ST1842/CC30 | PEN, MUP | blaZ, mup(A) | A | tsst-1 | ||
| SS | 3 | 1 | t352 | III | – | ST1842/CC30 | PEN, MUP | blaZ, mup(A) | A | tsst-1 | ||
| 5 | I | NS | 1/2 | 1/1 | t790 | I | – | ST22/CC22 | PEN | blaZ | B | cna |
| 6 | I | SS | 1 | 1 | t2155 | IV | – | ST121/CC121 | PEN | blaZ | B | cna |
| 7 | L | NS | 1/2 | 1/1 | t015 | I | – | ST45/CC45 | PEN | blaZ | B | cna |
| 8 | L | NS | 1 | 1 | t015 | I | – | ST45/CC45 | PEN | blaZ | B | cna |
| 9 | L | NS | 1 | 1 | t015 | I | – | ST45/CC45 | PEN | blaZ | B | cna |
Animal contact degree: H, high contact, continuous farm workers; I, intermediate contact, individuals living on the farm and occasional farm workers; L, low contact, individuals cohabiting with farm workers or with those living on the farm.
Periods of sampling: (1) time zero, April 2013; (2) at 6 months October 2013; (3) at 12 months, April 2014.
In relation to nasal carriage, 63.6% of individuals were S. aureus carriers. It is important to remark that all individuals that worked on the farm or that lived in the house in the farm (included in H or I groups depending on animal contact degree) were S. aureus carriers, either in the nose or in the skin. One of the farmers (case 2) carried the same clone of S. aureus along the three periods studied and was typed as t209/ST109/CC9.
Characteristics of the MRSA strain from the farmer carrierThe MRSA strain was recovered in the superficial skin lesion of one farm worker with high contact with animals. This strain was ascribed to spa-type-t843, agr-type-III, sequence-type ST130, clonal complex CC130 and SCCmec-XI. The strain showed susceptibility to all non-β-lactam antimicrobials and carried the mecC and blaZ-SCCmec-XI resistance genes, in addition to the virulence genes luk-DE and etd2. This strain lacked the genes of the IEC system.
Three nasal samples and two skin-lesion samples were obtained in the farmer who carried the mecC-MRSA strain. This mecC strain was recovered in one of the two skin samples analyzed, that corresponded to the third sampling period (during that period, the farmer was working in the field of traditional cheese production in a very close dairy cattle farm, besides working in the farm of this study). An additional S. aureus strain was also obtained in one of the three nasal samples of this farmer, but corresponded to a MSSA CC30 strain (Table 2).
Characteristics of the MSSA isolatesA high diversity of spa-types (n=9) was detected among the 17 MSSA isolates (t012, t015, t166, t209, t352, t790, t11719, t2155, t2678). Moreover, 9 different sequence types (ST22, ST30, ST34, ST45, ST109, ST121, ST133, ST1333, ST1842) belonging to 6 different clonal complexes (CC9, CC22, CC30, CC45, CC121, CC133) were identified in our study, being the most predominant STs/CCs being the following ones (% of strains): ST30 or ST34 or ST1333 or ST1842/CC30 (35.3%), ST45/CC45 (23.5%), and ST109/CC9 (17.6%). It is remarkable the detection of one S. aureus strain belonging to a lineage that has been associated with animals, as is the case of t2678/ST133/CC133. Interestingly, this strain was recovered from an individual who lived in the farm and worked sporadically on it. The most frequent agr-type detected was agr-I (7 strains) followed by agr-II (6 strains), agr-III (4 strains) and agr-IV (1 strain).
Antimicrobial susceptibility testing of MSSA strains showed the following results [% of strains/genes detected]: penicillin [100%/blaZ]; erythromycin [23.5%/msr(A), msr(B), mph(C), erm(A), erm(C)] and clindamycin [23.5%]; kanamycin [11.8%/aph(3′)-IIIa], tetracycline [5.9%/tet(K)], mupirocin [17.7%/mup(A)], streptomycin [5.9%,str] and fusidic acid [5.9%)/fus(C)]. All detailed results are shown in Table 2.
A high diversity of virulence factors was identified among the 17 MSSA strains (Table 2). Interestingly the 3 strains recovered from skin lesions of the same individual and typed as CC133 or CC30 contained the tsst-1 toxin gene, and 6 strains (isolated from three different individuals, case 1, 2 and 3) belonging to lineages CC9 or CC30 harbored the exfoliative toxin A gene (eta). The luk-DE genes were detected in 7 strains and the lukM gene was identified in the strain t2678/ST133/CC133. Eight strains of lineages CC22, CC30 and CC45 were positive for cna gene. All strains were negative for the presence of the genes lukF/lukS-PV, etb, etd and bap.
All MSSA strains harbored the genes of the IEC system except for two strains ascribed to ST133/CC133 and ST1333/CC30. The IEC type-B was predominant among the remaining strains (11 isolates), followed by IEC type-A (4 isolates).
DiscussionThe detection of a mecC positive MRSA strain from a skin lesion of one of the workers of the livestock farm is of great relevance, and the animal origin of this strain is highly suggested. The lack of the IEC cluster in this strain is of relevance, as it supports its potential animal origin. MRSA strains of the lineage CC130 have been detected in other studies in farm animals,17 mainly in cows and also in cow's milk.7 It is interesting to note that this farmer was working in the traditional production of cow cheese in a dairy farm close to the farm under study, where most animals were cattle. It is possible that the contact with the animals or with the milk could be in the origin of this strain. The characteristic profile of susceptibility for non-beta-lactams and the restricted virulence gene content to etd2 of our CC130 strain, agree with results of other mecC MRSA strains previously reported.18
The mecC-MRSA strain detected in our study was typed as t843-ST130-CC130, and it is one of the first cases described in farmers in Spain and the first one relating with cheese traditional making, with the potential risk that could entail its entry into the food chain. Some previous studies performed in Spain found the lineage t843/ST130 as the causative agent of fatal bacteremia in hospitalized patients or in a community patient.18–20 Other authors have identified similar genetic lineages (t1535/ST130) harboring mecC gene in wild small mammals14 or t843/ST2676 in urban wastewater,21 evidencing the spread of this lineage in different environments in Spain. Moreover, a case has recently been detected of S. aureus carrying mecC in livestock in Spain22 and in river water close to a livestock farm.23 Regarding the situation in other countries, MRSA strains typed as t843/ST130 have been described previously in humans in the European Union,10 but this lineage was also detected in different animal host species as cattle, cows, sheep or pets in UK, Denmark, Germany or France.10 Some authors suggested that MRSA mecC emerged in animals, mainly in ruminants, and subsequently made the leap to humans.9 In our case, the strain belonging to CC130 lacked the genes of the IEC system, and this fact points to an animal origin, probably being a potential case of zoonosis of LA-MRSA from cattle to human. In the same line, there are a few European studies with the objective of testing the possible zoonotic transmission between cattle and humans.24–26 In all of these studies, humans with different infections presented livestock animal contact or were linked to a farming activity. This fact supports the possible zoonotic transmission in our study.
Seven of the spa types identified in this study were ascribed to CC30, a lineage frequently found as colonizer of human microbiota,13,27 and it is also considered one of the most frequent S. aureus lineages detected in human infections.3 Strains of skin origin of lineage CC30, recovered from the individual of case 4, harbored the tsst-1 gene encoding the toxic shock syndrome toxin (TSST). It is interesting to remark that this person lived in the farm and often showed superficial and difficult healing wounds. The presence of relevant virulence factors (typical of community-associated S. aureus) and the low content in resis-tance determinants have been previously reported by other authors in MRSA-CC30 strains.28 Interestingly one MSSA strain typed as t11719-ST1333-CC30 showed a multidrug-resistance phenotype (penicillin-erythromycin-clindamycin-tetracycline-fusidic acid-mupirocin) with the detection of several resistance determinants (Table 2). This genetic lineage has been previously reported in canine samples from Japan29 and the absence of the genes of IEC system points to a possible animal origin of the strain.13
In the same line, detection of one MSSA strain typed as t2678-ST133-CC133 is remarkable because in previous studies it has been detected in small ruminants such as goats or sheep.30 The detection of gene lukM, prevalent in ungulates, ruminants and bovine mastitis,7 and the absence of IEC genes, supports the idea of the possible animal origin of the strain. Interestingly, some authors have suggested a possible ancestral human origin of the genetic lineage CC133 and have hypothesized about the possibility of jumping to ruminants few centuries ago.31
MSSA strains detected in five members of the family (belonging to animal-contact degree groups I and L) were assigned to hospital-associated clonal complexes CC22, CC45 and CC121.4 Therefore, some authors have postulated about the ability of MSSA to serve as recipient of the SCCmec cassette, subsequently losing any of its components, as the mecA gene.27 Moreover, the lineage t2155-ST121-CC121 found in one of our MSSA strains, has also been detected among MRSA implicated in skin and soft tissues infections.32 Interestingly, this strain was isolated from a skin lesion of an individual who worked sporadically in the farm (Case 6). In the same line, many studies have associated the MRSA-ST22 genetic lineage with frequent infections in Europe. This lineage in its methicillin-resistance form was called EMRSA-15.33 In our case methicillin-susceptible CC22 clade was detected.
As for the single case (case 2) where colonization was maintained by the same CC9 clone of S. aureus over the period studied, it is interesting to note that this genetic lineage was initially associated with farm animals, mainly pigs in Asia.34 Similarly CC9 has been detected in meat samples in Spain35 and in bloodstream infections in France,36 with similar patterns of resistance, particularly resistance to erythromycin, but the difference is of methicillin resistance in our case. This MSSA profile and the presence of IEC type-B, points to the possible human origin of this clone and reveals its great capacity for colonization in different environments. Furthermore, the presence of the gene encoding the exfoliatin A (eta) on the CC9 strains of this individual is noteworthy. Previous studies have suggested a high association between the CC9 and its ability to produce exfoliative toxins.27
ConclusionsWe report the detection of the lineage ST130/CC130 containing the novel methicillin-resistance-determinant mecC as well as other livestock-associated S. aureus lineages (as CC133 or CC9) in nasal or skin samples of a family of livestock farmers in Spain. The mecC-CC130 strain was isolated from the superficial skin lesion of a farmer which had daily contact with farm animals, mainly cattle, and with traditional cheese production. Cows have been postulated as the main reservoir of this emerging genetic lineage. This fact highlights the zoonotic potential of this emergent resistant microorganism and other similar animal-related clades, with the risk that this entails for human health. More studies should be performed in the future to assess the real risk for colonization and infection of farmers in relation to MRSA-mecC clones and the different reservoirs that may exist in nature.
Conflict of interestThe authors declare no conflict of interest.
This project was supported by project SAF2012-35474 from the Ministerio de Economía y Competitividad of Spain and the Fondo Europeo de Desarrollo Regional (FEDER). Daniel Benito has a predoctoral fellowship from the Ministerio of Economia y Competitividad of Spain (reference BES-2010-039341) and Paula Gómez has a predoctoral fellowship of the University of La Rioja, Spain (reference). Carmen Lozano has a contract associated with project SAF2012-35474.