A multiresistant CTX-M-15-producing clonal group of Escherichia coli isolates, namely O25b:H4/ST131, has recently emerged in three continents. At this moment, appropriate studies to assess the real prevalence of this successful lineage are still scarce.
MethodsIn a prospective study in the south of Spain, among all clinical E. coli isolates recovered in Seville during a 30 week period in 2010, ST131 was screened by using PCR for O25b/pabB3/B23 traits. ESBL enzymes were characterized by PCR and sequencing. Genetic relatedness was performed by XbaI PFGE.
ResultsThis clonal group was found to be prevalent (12.5% of all E. coli isolates), and only 37 (6.8% of ST131 isolates) were ESBL producers. Among 25 characterized ESBL-producing ST131 isolates, 96% harbored CTX-M-15. ST131 isolates were more frequently resistant to amoxicillin/clavulanate, aminoglycosides and fluoroquinolones in both ESBL and non-ESBL producers groups. XbaI PFGE performed on 88 ST131 isolates showed three pulsotypes, which included ≥4 isolates each (25% of all typed ST131 isolates), and 11 pulsotypes, which contained 2–3 isolates each. Three of 14 pulsotypes of this clonal group included both nalidixic acid-resistant and susceptible isolates, and five pulsotypes included both ESBL and non-ESBL producers.
ConclusionsOur findings suggest that O25b/ST131 is a prevalent clone in our area, and the observed prevalence of ESBL-producers within this clone is similar to that found in the total isolates of this species. Certain pulsotypes among ST131 clone that showed a greater expansion, and ESBL genes acquisition or quinolone resistance could explain part of this prevalence.
Recientemente, y de forma simultánea en 3 continentes, ha emergido un grupo clonal de E. coli multirresistente y productor de CTX-M-15. Por el momento, se dispone de pocos estudios que analicen de forma apropiada la prevalencia real de este linaje.
MétodosEn un estudio prospectivo en el sur de España, realizado con todos los aislados clínicos de E. coli recuperados en Sevilla durante 30 semanas en 2010, se realizó el despistaje de ST131 mediante PCR para O25b/pabB3/B23. Los enzimas BLEE fueron caracterizados mediante PCR y posterior secuenciación. La relación genética de los aislados fue estudiada mediante PFGE con XbaI.
Resultadosla prevalencia de este grupo clonal resultó ser del 12,5% de todos los aislados de E. coli y únicamente 37 (6,8% de los aislados ST131) eran productores de BLEE. Se caracterizaron 25 aislados ST131 productores de BLEE y la mayoría (96%) producían CTX-M-15. Los aislados ST131 eran con más frecuencia resistentes a amoxicilina/clavulánico, aminoglicósidos y fluorquinolonas tanto en el grupo de productores de BLEE como en el de no productores. En el análisis mediante XbaI PFGE, realizado a 88 aislados ST131, se observaron 3 pulsotipos, que incluían ≥4 aislados cada uno (25% de todos los aislados ST131 tipados) y 11 pulsotipos, que contenían 2-3 aislados cada uno. Tres de los 14 pulsotipos de este grupo clonal incluían aislados sensibles y resistentes al ácido nalidíxico y 5 pulsotipos incluían productores y no productores de BLEE.
ConclusionesLos hallazgos de este estudio sugieren que O25b/ST131 es un clon prevalente en nuestra área y la prevalencia de BLEE en el referido clon es idéntica a la que se encuentra en la totalidad de los aislados de la especie. Algunos pulsotipos dentro del clon muestran una expansión mayor que podría ser explicada en parte por la adquisición de genes codificantes de BLEEs o resistencia a quinolonas.
A multiresistant CTX-M-15-producing clonal group of Escherichia coli isolates, namely O25b:H4/ST131, has recently emerged and spread in nosocomial and community settings across three continents,1 including our area. Isolates belonging to this clone present a significant number of virulence factors, and, in addition to the multiresistance pattern shown worldwide, which includes determinants such as blaCTX-M-15, blaOXA-1, blaTEM-1, aac (3)-II and aac (6′)-Ib-cr, susceptible non-ESBL-producing isolates have been also detected.2 Whether the ST131 clone came from a successful lineage before they acquired multiresistance-encoding plasmids or chromosomal quinolone resistance is not well known.3 At the present time, scarce data of the actual prevalence of this successful lineage are available. A nacional multicenter study has been carried out in 2009 in Spain, where ST131 proved to be the most prevalent clone, accounting for 12% of isolates overall.4 This study was conducted during one month and were not available so the information throughout the year. We report here on the prevalence of ST131 among all E. coli isolates in our area during 2010.
Material and methodsAll E. coli isolated at the Virgen Macarena and the Virgen del Rocío University Hospitals, Seville, Spain, during 30 weeks in 2010 were included. The Clinical Microbiology laboratories in these hospitals receive samples from primary care patients, ambulatory patients receiving specialist attention, and admitted patients (two tertiary hospitals and two long term care hospitals), in an area with 1 million inhabitants. Only the first isolate per patient was included. All isolates were screened for belonging to the O25b:H4/ST131 clonal group, using PCR with primers for O25b rfb and allele 3 of the pabB gene,5 and multiplex PCR for phylogroup B23 typing.6 Microscan panels were used for susceptibility testing. ESBL production was screened by double-disk synergy test. One-hundred forty-two ESBL producers were further analyzed and enzyme type was determined by PCR assay, using primer sets for the ESBL groups (CTXM, SHV and TEM) and sequencing. An isolate was defined as non susceptible when it was resistant or intermediate according to the Clinical and Laboratory Standards Institute breakpoints.7 Comparison of proportions was performed by contingency table using the chi-squared test, and the Fisher's exact test when the expected values in any of the cells were below 10.
Genetic relatedness of the first 88 O25b:H4/ST131 positive isolates and the first 47 non-ST131 ESBL producers was performed by XbaI PFGE analysis (http://www.pulsenetinternational.org/protocols/pfge.asp). Dendograms were created with Fingerprinting 3.0 software (BioRad), using the Dice coefficient and a position tolerance of 1%. Isolates exhibiting <3-band difference (corresponding to >94% similarity) were assigned to the same pulsotype.
ResultsA total of 4308 E. coli isolates were analyzed during the study period. These were recovered from urine samples (88.9%), wounds and abscesses (6.0%), the bloodstream (3.9%), the respiratory tract (1.0%), and other sources (0.2%). Five hundred and forty (12.5%) cases were found to be positive for O25b/pabB3/B23. Thirty-seven (6.8%) ST131 isolates were ESBL producers, and the remaining 503 (93%) ST131 isolates were non-ESBL producers. The overall prevalence of ESBL producers among all E. coli isolates was 6.8% (295 isolates). Of the 142 ESBL genes analyzed, 52 (36.6%) corresponded to CTX-M group 1 (CTX-M-15, 43 isolates; CTX-M-1, 5 isolates; CTX-M-32, 4 isolates), 48 (33.8%) to CTX-M group 9 (CTX-M-14, 41 isolates; CTX-M-9, 5 isolates; CTX-M-27, 2 isolates) and 42 (29.6%) to SHV group (SHV-12, 41 isolates and SHV-8, 1 isolate). Most ESBL producers belonged to phylogroup A (32%) and B1 (30%); whereas 43 (30%) isolates belonged to phylogroup B2 (25 of which corresponded to the ST131 clone). Twenty-four of 25 (96%) ESBL-producing ST131 isolates coded for CTX-M-15 enzyme and one for CTX-M-14.
Among non-ESBL producers, ST131 isolates (as compared to non-ST131 isolates) were significantly more frequently non susceptible to amoxicillin/clavulanate (27.6 vs. 18.0%, p<0.01), gentamicin (17.3 vs. 7.7%, p<0.01), tobramicina (20.2 vs. 7.9%, p<0.01), amikacin (1.2% vs. 0.1%, p=0.006), nalidixic acid (76.1 vs. 40.9%, p<0.01) and ciprofloxacin (72.8 vs. 32.6%, p<0.01). Similarly, in the ESBL subgroup, ST131 isolates were more frequently non susceptible to amoxicillin clavulanate (82.4 vs. 28.6%, p<0.01), tobramycin (80.0 vs. 19.9%, p<0.001), amikacin (28.0 vs. 1.3%, p<0.01) and ciprofloxacin (97.1 vs. 81.5%, p=0.022) than other ESBL producers. No differences were found between ST131 and non ST131 isolates for resistance to co-trimoxazole or fosfomycin.
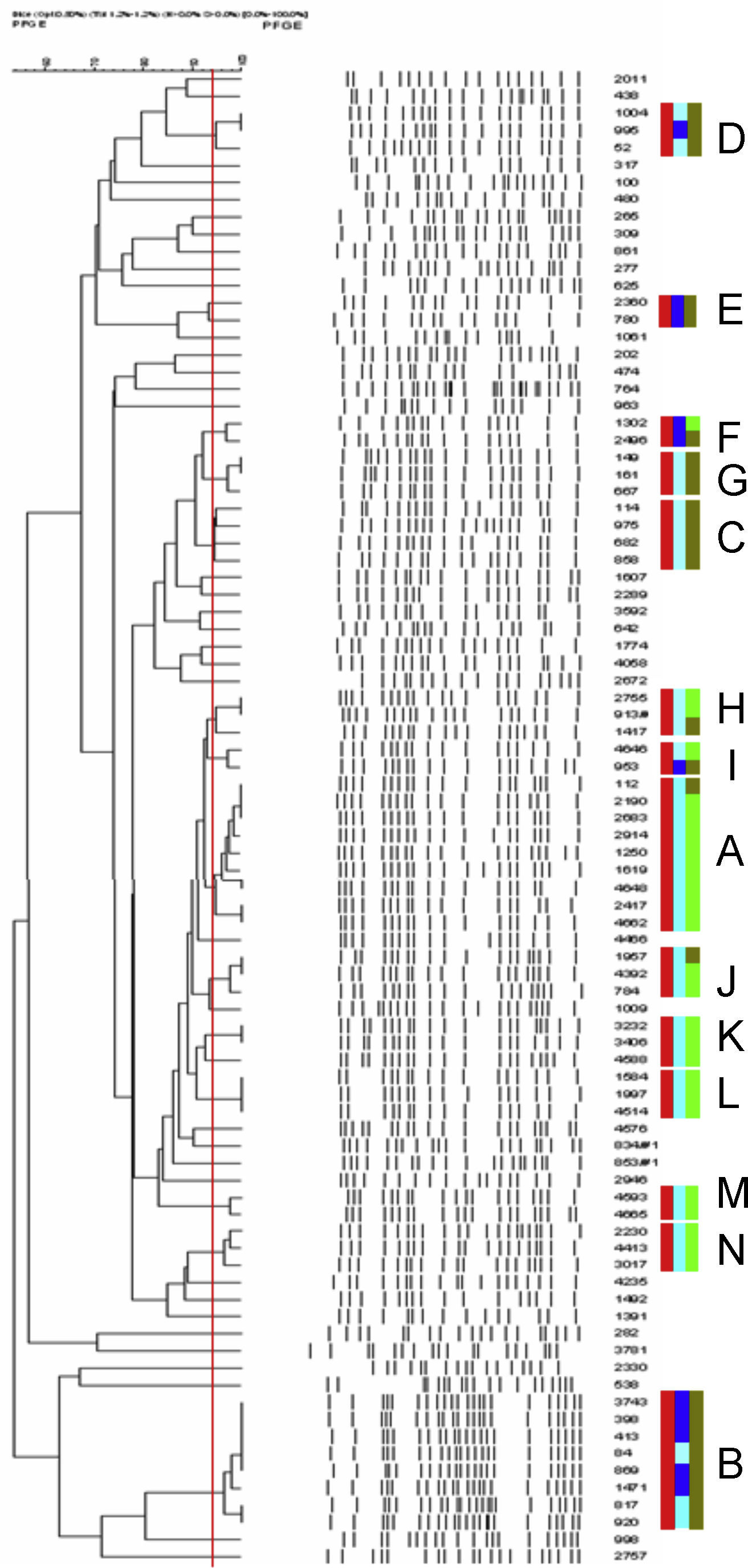
Eighty-eight ST131 isolates yielded 50 distinct pulsotypes: one pulsotype (A) contained 10 isolates, one (B) contained 8 isolates, one (C) contained 4 isolates, eleven pulsotypes (D–N) contained 2–3 isolates and 37 single-isolates pulsotypes (Fig. 1). On the other hand, 47 non-ST131 ESBL producers yielded 42 distinct pulsotypes: two pulsotypes contained 3 isolates, one contained 2 isolates and 39 single-isolates pulsotypes. Pulsotypes of ST131 containing more than 1 isolate included 68% of typed CTX-M-15-producing isolates. Three pulsotypes among the ST131 isolates included both nalidixic acid-resistant and susceptible isolates and five pulsotypes included both ESBL producers and non-producers isolates.
Dendogram of XbaI PFGE patterns of 88 O25b/pabB3/B23-positive isolates. Red rectangles grouped isolates belonging to same pulsotype (<3-bands difference, Dice similarity value >94%, red line). Light blue rectangles correspond to quinolone resistant isolates and strong blue rectangles to quinolones susceptible. Light green rectangles correspond to ESBL producers and strong green rectangles to non-ESBL producers. Capital letters A–N were used to design pulsotypes contained >1 isolate. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
Our findings showed that ST131 is currently a predominant clone in our area and that the majority of the isolates lacked ESBL genes. The frequency found in our prospective surveillance study correlated with estimates calculated by Johnson et al. from data obtained from selected mostly invasive isolates3 and the prevalence found in the Spanish multicenter survey.4 Like them, we also found that aminoglycoside and quinolone-resistance were more frequent in ST131 isolates. The fact that different ST131 pulsotypes included both nalidixic acid-resistant and susceptible isolates is a new finding and contrasts with the hypothesis that current fluoroquinolone-resistant ST131 isolates derive from a recent common fluoroquinolone-resistant ST131 ancestor.3 Johnson et al.,3 failed to find quinolone-susceptible isolates among those belonging to the ST131 clonal group. This may have been due to local epidemiological differences, or to the fact that most of the isolates in their study were obtained from blood cultures and other invasive infections, while most of ours were isolated from urine.
Certain pulsotypes within ST131 group shown wider expansion, some of them carrying CTX-M-15 but it can be seen also a group of 8 isolates, 3 of them nalidixic acid resistant and MIC values for ciprofloxacin >1mg/L, without ESBL production. In an analysis of a collection of 579 diverse E. coli ST131 isolates, also a small number of high-frequency pulsotipes were found predominant both in Canada and U.S.A. and other countries.8 In this recent survey, resistance traits as fluorquinolone resistance or ESBL production seemed to be associated specifically with some pulsotypes. Our results are in part consistent with these findings. Most of our ST131 ESBL-producers harbored CTX-M-15 enzyme and were grouped in seven pulsotypes, which 4 contained both ESBL-producers and non-producers. This finding suggests the horizontal spread of this gene by one vector or multiple related plasmids. BlaCTX-M-15 gene has been mainly found in diverse Inc FII and FIA plasmids in E. coli isolates belonging to this clone (revised in reference 9), but also this determinant has been detected in other plasmids as Inc A/C in a hospital in Madrid, where Inc FII plasmids were found predominant.10 Others CTX-M enzymes, as CTX-M-14, have been less frequently associated to ST131 in Spain,11 in concordance with our results. In contrast, in some Asian surveys CTX-M-14 was the most common enzyme associated to Inc FII and Inc I1 plasmids.12 When ST131 isolates from different origins have been studied, diverse Inc FII plasmids were detected.13 In order to know if an epidemic or multiple plasmid vectors are implied in the local spread of ESBL genes within ST131 clone in our area, a further analysis is required. Multiple factors, in combination with quinolone resistance or blaCTX-M-15, could be contributing to the success of ST131, especially in the case of some pulsotypes, in our area during 2010.
FundingThis work was partially supported by the Ministerio de Ciencia e Innovación (Instituto de Salud Carlos III, Fondo de Investigación Sanitaria, REIPI- RD06/0008-1018, PS09/01273, PI10/01955 and PI070190), the Ministerio de Educación y Ciencia (AGL-2008-02129), Junta de Andalucía (PI-0048/2008, P09-CTS-5259, PI 0034-2009).
Conflict of interestThe authors declare no conflict of interest of any nature.