The use of new technologies, such as proteomics, permits the efficient and reproducible analysis, identification and characterisation of peptides and proteins from different biological and non-biological matrices. This can help in the development of new biomarkers in the forensic sciences. Protein markers are highly resistant to the passage of time and adverse environmental conditions, and could provide a broad overview of the physiological status of the subjects. The use of protein markers substantially reduces the contamination of samples as compared to DNA, while providing quantitative and highly reliable data that are backed by databases for their interpretation. This work presents a review of the advances and limitations of proteomics to establish the origin of the evidence found, the cause of death, the presence of pathogens and conditional disease, as well as the biological age at death, the post-mortem interval, or the biogeographic origin of subjects.
El uso de nuevas tecnologías como la proteómica permite analizar, identificar y caracterizar péptidos y proteínas provenientes de diversas matrices biológicas y no biológicas, de forma eficiente y reproducible. Esto puede facilitar el desarrollo de nuevos biomarcadores en el área de las ciencias forenses. Los marcadores proteicos son altamente resistentes al paso del tiempo, condiciones ambientales adversas y proporcionan un amplio panorama del estatus fisiológico de los sujetos. El uso de marcadores proteicos reduce sustancialmente la contaminación de las muestras en comparación con el ADN, al tiempo que aporta datos cuantitativos y confiables, que se encuentran respaldados por bases de datos para su interpretación. En este trabajo se hace una revisión de los avances y limitaciones de la proteómica para establecer el origen de los indicios hallados, la causa de muerte, la presencia de patógenos y enfermedades condicionantes o la edad biológica al momento del deceso, el intervalo post-mortem o el origen biogeográfico de un individuo.
The development in the separation, detection and identification of peptides and proteins using techniques such as: one and 2 dimensional electrophoresis, immunodetection by means of multiplex ELISA, high performance liquid chromatography (HPLC/UHPLC) and mass spectrometry; added to efficient bioinformatics analysis, which allows the organisation and management of large amounts of information, have honed proteomics into a valuable tool in the areas of clinical and biotechnological research.1,2 Currently, the high quality of this large amount of information has enabled the development of new pharmacological strategies, the identification of pathogens, the discovery of new therapeutic targets, the optimisation of bioindustrial processes, and the implementation of ground-breaking techniques in disease detection and prevention.3–5 Proteomics as a biological problem-solving strategy presents a high degree of flexibility and reproducibility due to the possibility of obtaining data from a wide variety of matrices such as tissue, blood, saliva, semen and hair, among others.6–8 At the same time, the techniques used to obtain and purify proteins and peptides are highly compatible with the analysis of traces of other elements such as heavy metals, accelerants and solvents, and traces of explosives or toxins, and this makes it a powerful tool in the forensic context.9,10
In the forensic area, much of the physical information that could be provided by biological evidence depends on the quantity and quality of the sample obtained, and on the efficiency of identification and analysis techniques used by specialist personnel. Unfortunately, despite the efforts of experts and forensic personnel, in these contexts the preservation and abundance of biological evidence is not always optimal, which hinders biochemical analysis and obtaining results with a high degree of scientific proof in the legal process.10–13
Although the gold standard for identifying individuals in a forensic context is based on the amplification of fragments of DNA, the study of proteins in the forensic context is a new and powerful tool that can overcome some of the shortcomings of the conventional techniques since, unlike nucleic acids, proteins are molecules that are highly stable in physical and environmental conditions, and preserve well for prolonged periods.14,15 Proteomics also provides information on the physiological status of individuals, the presence of pathogens, exposure to toxic substances and even enables the identification of specimens of high biological value in the forensic context, such as insects or plants.16–20
Thus, all these characteristics make proteomics an emerging tool of great potential in forensic science. Throughout this review we will address the most relevant advances that have been made in various proteomic studies in the forensic area, and indicate some of the limitations that might be encountered when using this sample analysis strategy.
Proteomics as a source of biological information in the context of forensicsThe high specificity in proteomic detection and analysis provides a great variety of information on different aspects, such as the species to which a certain sample belongs, an individual's state of health and even, by means of bioindicators, environmental conditions.1,9,21,22
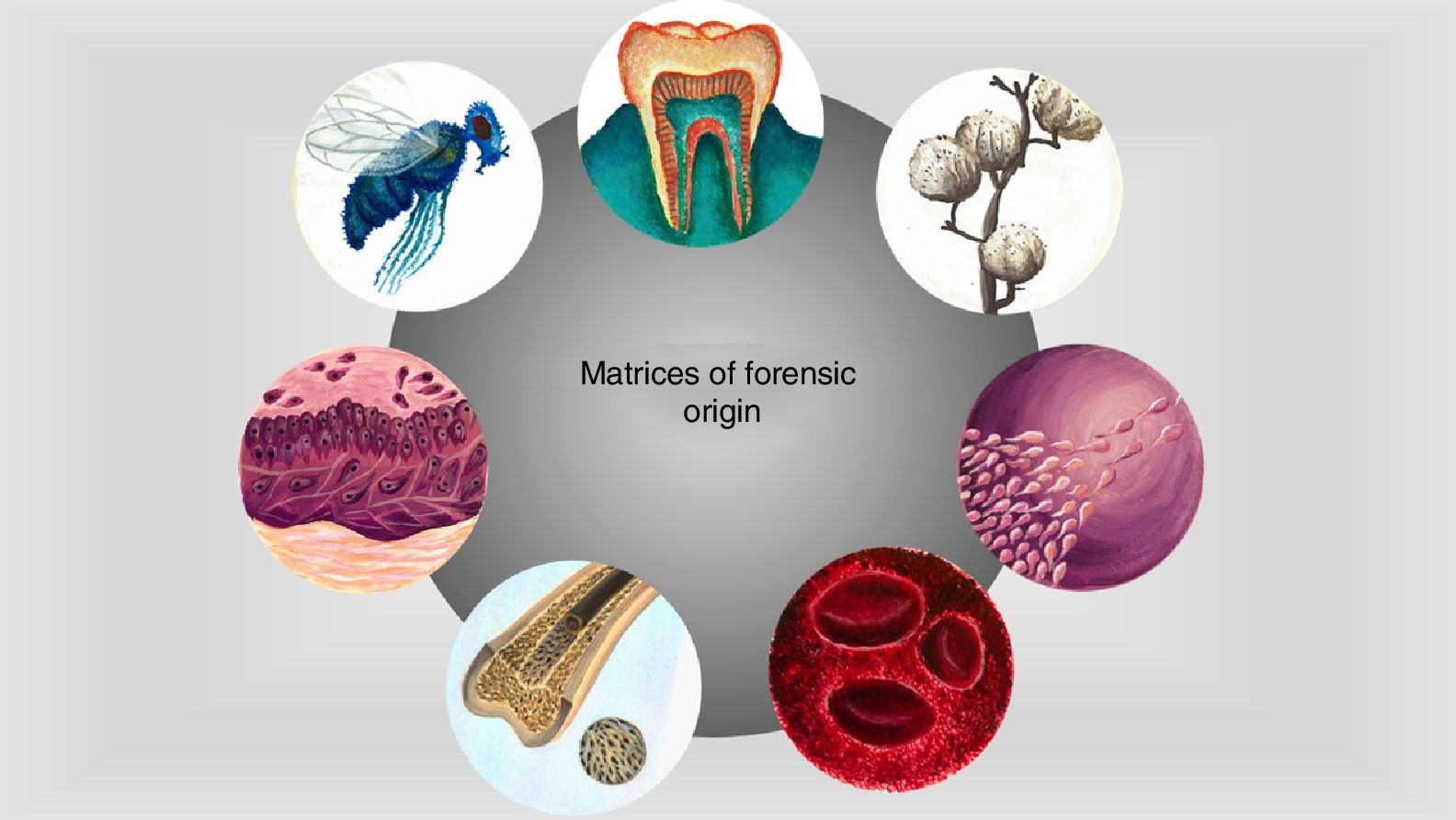
The area where the forensic investigation takes place is made up of vestiges of events relating to accidents, crimes, natural disasters or armed conflict. Although these vestiges vary in nature, they are all characterised by being fragile and labile under environmental conditions.23–25 There are endless possible pieces of material evidence that can be observed in legal proceedings, ranging from fingerprints to body fluids, and they are key pieces in the development of an investigation, and in the reconstruction of events that have occurred in the past.25,26 Much of the evidence that can be available for study is of biological (blood, skin fragments, fluids) or non-biological origin (textile fibres or soils) (Fig. 1), which contain protein molecules that, after isolation and characterisation, can provide information that goes beyond human identification or determination of ancestry.10
Sources of protein information commonly found at the forensic investigation site. Matrices of biological origin such as teeth, plants, insects, biological fluids, bone tissue or skin fragments contain proteins that can provide information on the physiological status of individuals, their ethnic origin and even their biological sex. This information can be isolated and analyzed by proteomic strategies providing evidence in the forensic context (original images).
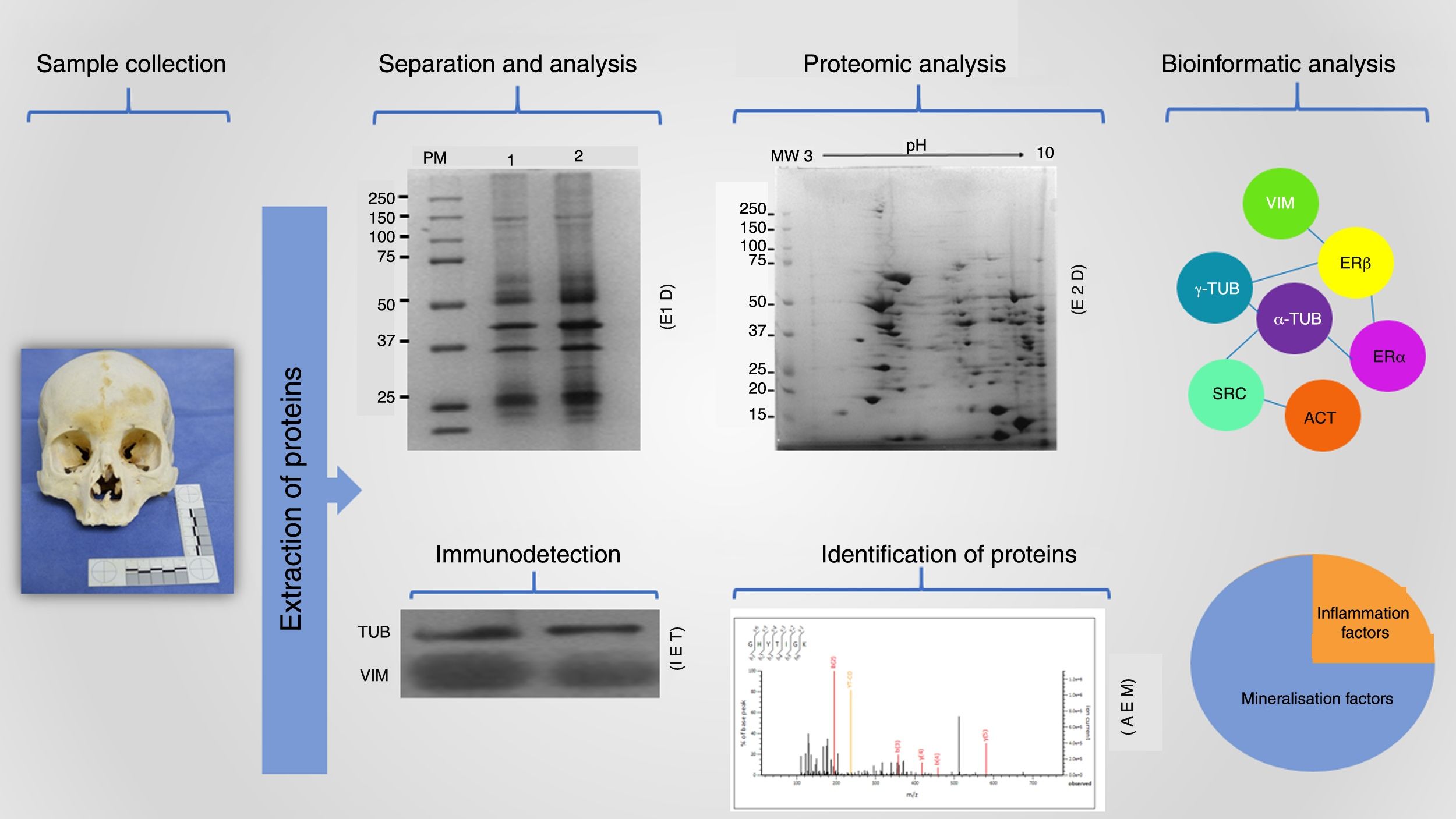
The detection of protein markers is crucial for resolving various questions that can arise in the forensic area; this detection and characterisation is done following a highly reproducible workflow (Fig. 2). This starts with collecting the sample, which must be under optimal conditions, followed by the extraction of the proteins present using an appropriate strategy for each type of matrix found at the investigation site. The procedure must allow protein molecules to be obtained in sufficient concentrations for separation and analysis. Likewise, these isolated molecules can be identified by means of mass spectrometry coupled with high resolution liquid chromatography, which, with the analysis of such small volumes (of up to 10μl) provides a large amount of data that are processed by means of bioinformatic tools that enable the information to be filtered and categorised to identify protein markers that are of value in the forensic field.2,17,19,22
Proteomics workflow in the forensic context. Proteins extracted from a matrix obtained at the forensic investigation site are isolated by means of different techniques, such as one- and two-dimensional electrophoresis (1DE and 2DE respectively). Also, these molecules can be identified using immunodetection strategies such as immunoelectrotransference (IET) or through the identification of peptides and proteins by mass spectrometry analysis (MSA). All these data together with a robust bioinformatics analysis provide accurate and highly reproducible information (original images).
Since metabolic indicators of some biological events remain present in the body after a traumatic event, it is possible to identify markers specific to each type of event. For example, by isoelectric point electrophoresis it is possible to differentiate substances of abuse and doping, such as recombinant erythropoietin, from their endogenous analogues. This is a glycoprotein hormone that stimulates erythropoiesis by increasing the performance of professional athletes. During doping detection tests, the use of proteomic strategies such as isoelectric point electrophoresis allows the detection and differentiation of recombinant protein from endogenous protein in a quantitative and precise manner, because the recombinant protein may have a particular electrophoretic profile, depending on the cell line in which it was produced and the purification method used.27
In this regard, the consumption of other more common substances of abuse leaves metabolic evidence that can be traced by proteomic analyses. Either in post-mortem tissue, as reported in alcoholism, when the protein profile of nerve tissue in the cerebral cortex is modified,28 or with samples that are more easily obtained, such as serum, where it is possible to detect modifications in haptoglobin and thioredoxin levels in subjects addicted to heroin.29
Another type of crucial biological information in the forensic context is determining the biological age of remains found. Normally, these parameters are determined based on a morphological analysis performed by an anthropologist or forensic dentist, which can make them difficult to quantify precisely. The use of new strategies, such as proteomic analysis of bone remains, can provide a new set of biomarkers for more reliable estimation of biological age. It has been demonstrated recently that there is a correlation between the abundance of fetuin A present in bone tissue and the biological age of individuals. This suggests that it may be a new marker for estimating the biological age of bone remains linked to a forensic context.30 This provides quantitative tools for resolving these types of problems and opens the door to new strategies for determining the age of individual remains by highly reproducible techniques that are supported by robust databases for their interpretation.
Proteomic analysis of blood holds valuable information in the forensic sceneIn the forensic context, blood represents one of the most important pieces of biological evidence, since it is one of the main indicators in a violent event and difficult to eliminate from the forensic investigation site, and therefore traces can last for relatively long periods of time. In addition, this fluid stores crucial biological information regarding the victim and/or the offender, such as their sex, blood type or toxicological condition, among others.31
Given the enormous potential of this type of sample from a legal perspective, the first step in using blood as forensic evidence is to identify it correctly, as contamination by particles, chemical substances or other biological fluids such as saliva, semen or blood from other species is common in this context.32,33
Since each biological fluid has a specific function, the nature and concentration of proteins present in a specific fluid is unique, and can serve as a signature to distinguish one fluid from another. Using proteomic strategies, fluid markers have been identified in blood or other fluids from small amounts of sample, which suggests that it is possible to identify fluid-specific markers, such as amylase in saliva, prostatic antigen in semen or β-spectrine and haemoglobin in the case of blood. Unlike other techniques, the use of strategies such as mass spectrometry allows precise identification of the various protein markers, reducing the risk of cross-identification, while even enabling the identification of multiple markers.34,35
In the forensic context, the presence of multiple fluids with diverse origins is not uncommon10,36; therefore, determining the origin of the sample, the species and even the condition in which it arrived at the forensic investigation site can be as important as the full identification of blood. Although there are many methods that enable identification of blood stains in situ, these have the disadvantage of being non-specific in determining the origin of the blood, while some are very destructive and not very compatible with the preservation of valuable biological information.26
A clear example of this is the identification of menstrual blood, which must be differentiated from circulatory blood in the case of a sexual assault.37 Although the use of markers such as RNA have proved an efficient technique in identifying fluids such as semen or circulatory blood, the identification of RNA markers from other fluids, such as saliva or menstrual blood, is still in its incipiency.38,39 In contrast, recently, some studies based on identifying proteins in menstrual blood by means of mass spectrometry have succeeded in determining many characteristic markers of this fluid, such as orexin-A (OREX) and the semaphorin receptor (PLXA1, D1), which are involved in the formation of umbilical cord vascular tissue and adhesion to the extracellular matrix.22,40
During forensic investigation, another common problem is identifying the species to which the blood spots found belong. Although the advent of molecular biology technologies has resulted in the identification of mitochondrial DNA fragments, called genetic barcodes, which enable the identification of the species in samples of uncertain or unknown origin in a highly efficient manner,41 this technology is still susceptible to sample degradation under unfavourable environmental conditions, and analysing old samples is still complex.42 In this regard, it has been demonstrated that by means of reverse-phase high performance liquid chromatography (RP-HPLC) it is possible to identify the species to which a blood sample or spot belongs from the chromatographic profile of the haem group, present in the haemoglobin. This technique has even enabled classification at species level in relatively old samples. This method, compared to other identification techniques, is based on a simple, highly efficient method of broad sensitivity and reproducibility, which could make it a valuable identification tool.43,44
Since blood contains amino acids, proteins, hormones and blood cells, among other things, it provides valuable information regarding the metabolic status of subjects, the presence of diseases and even the influence of psychoactive substances or toxins.25,45,46 This physiological status of individuals, whether victims or aggressors, can shed light on the events that occurred or on the presence of conditioning diseases, such as mental or neurodegenerative disease,21,47 which, in some cases, have been shown to be associated with violent scenarios or road traffic accidents.48,49 These types of disease are difficult to detect, and in forensic scenarios are generally diagnosed post-mortem through histological analysis. Thanks to advances in immunoproteomics and metabolomics, it has been suggested that slight changes in hormonal and metabolic markers could be detected in blood, using the multiplex ELISA technique, in live subjects in order to make a differential diagnosis in mental diseases such as schizophrenia or bipolar syndrome.50,51
As has been shown, blood is very valuable as evidence in the forensic context. Unfortunately, much of the biological information it contains can lose its legal validity due to contamination, degradation, or mishandling of the sample. It is therefore possible that the implementation of new analysis technologies, such as those included in proteomics, will overcome these barriers and provide information of high biological value and validity from a legal perspective on the origin of the samples or the metabolic and health status of the victims and/or aggressors, since the techniques used for protein analysis are highly reliable, efficient and reproducible.
Proteomic study of ancient and contemporary skeletal remains can reveal key forensic and anthropological informationMuch of the biological evidence found in the forensic context is housed within tissues such as skin, muscle, hair or in fluids such as saliva, semen and blood, among others, but very often the evidence contained in these tissues and fluids can be lost through biological events related to soft tissue decomposition.10,35 Therefore, it is common in forensic scenarios for bone tissue to represent most of the biological evidence found, since it preserves well over long periods, harbouring good quality biological information, both in forensic scenes and in sites of archaeological interest.52,53
This tissue is highly specialised and undergoes relatively slow replacement inside the organism. Its constituent cells regulate the dynamics of a highly mineralised extracellular matrix composed mostly of hydroxyapatite, glucans and associated proteins.52 As a whole, both the cells and the mineralised matrix participate in different events of metabolism, such as pH regulation, mineral and trace element metabolism, while being intimately associated with the endocrine and renal systems, and the metabolism of sugars and lipids. Therefore, bone tissue can store information on the present and past physiological status of individuals.54,55
Recently, the characterization of protein markers in bone tissue been shown to provide valuable information for resolving key questions from the forensic field, such as the presence of diseases, conditions of stress, habits and/or behaviours and even determining the post-mortem interval or the biological age of an individual. Added to this, by mass spectrometry, it is possible to detect spontaneous post-translational modifications (PTM) suffered by proteins, which are directly associated with the biological history of the sample up to time that it is analysed.6,30,52,53,56–58
The proteins contained in bone tissue are efficiently preserved with a high degree of conservation, to the extent that it is possible to analyse proteins as old as those isolated from the femur of a 43,000 year-old mammoth (Mammuthus primigenius), enabling the identification of molecules such as albumin, and the characterisation of various changes related to oxidation and post-mortem damage in the proteins.59
Thanks to this capacity of proteins to last over time under extreme environmental conditions inside bone tissue, it is possible to isolate and characterise specific markers to study the microbiome present in bone, which enables clinically important pathogens to be detected. Recently the presence of peptides from Mycobacterium protein markers has been identified as subunit A of fumarate reductase and adenylate kinase, by mass spectrometry, in bone remains from Roman settlements dating back to the 2nd and 4th centuries.60 Likewise, the protein AG85, one of the main secretion antigens produced by Mycobacterium tuberculosis, has been detected through the use of antibodies in bones from the middle-ages, allowing us to trace for the presence of this disease in these populations.61
Bone remains commonly show clear evidence of wear and tear due to physical activity, trauma or even violence.62 Therefore, the search for markers of bone tissue damage and repair processes can help clarify events that have occurred in both legal and anthropological contexts. Since mechanical events cause changes in the expression of genes and proteins in bone tissue, molecular markers can be found that evidence the presence of stress or mechanical fatigue, these markers, generally related to pro- and anti-apoptotic factors, may represent clear signs of trauma or wear and tear form physical activity.63,64 Likewise, some diseases such as osteoarthritis or other degenerative disease related to bone tissue can leave markers of damage and repair,65,66 which from a forensic context could be useful in determining the nature of lesions found in bone remains.66,67
In scenarios of decomposition and skeletonisation of individuals, determining the post-mortem interval may be difficult during a forensic investigation, although different methods and formulae have been developed to establish this interval; these are subject to conditions such as the weight of the individual, their age, the presence of disease, as well as the environmental conditions of the place of the events.68 Another strategy for calculating the post-mortem interval is the analysis of the succession of insects that over short and long periods has proved highly effective.69
From the molecular approach it has been possible to determine through immunohistochemistry and messenger RNA analysis that the hypoxia-inducible factor present in gingival tissue can be a suitable marker for calculating this interval in short periods, but both strategies can be ineffective in cases of very prolonged periods of decomposition.70
It has recently been demonstrated that proteomic analysis of bone remains in an advanced state of decomposition and skeletalisation can enable the identification of markers that help determine this interval precisely.58 Different proteins, such as haemoglobin or serum transferrin, show a marked reduction in their relative abundance in periods of up to 6 months after death, while the relative abundance of other proteins such as albumin or apolipoprotain A1 remains constant over the same period of time. Similarly, the desamination of biglycan, a leucine-rich repeat proteoglycan presents a positive correlation with the post-mortem interval, which outlines this molecule as a potential biomarker for determining this interval.58
Together these antecedents indicate that proteomic analysis of the bone matrix can provide crucial information for identifying markers that account for pre-mortem events such as trauma, habits and behaviours, infectious diseases and even in resolving questions such as the post-mortem interval in skeletalised remains.
Nearby applications in the forensic field: characterisation of polymorphic peptides as an auxiliary identification toolAlthough the use of fingerprints is the best and most common way of identifying individuals, events such as calcination or prolonged periods of decomposition can make the process difficult. Therefore, generally in the forensic context, identification under these circumstances can limit the progress of the investigation due to the loss of the physiognomic characteristics that enable identification. Although it is possible to complete the identification process based on other physical characteristics, such as the organisation and morphological characteristics of the teeth, this approach has certain limitations, since it requires comparison with clinical records or with some type of pattern to enable analysis of the evidence found in order to carry out a full identification.71,72
At present, molecular tools based on amplifying and identifying DNA fragments, such as the characterisation of single nucleotide polymorphisms, the characterisation of repeated “microsatellite” sequences, and the identification of mitochondrial DNA holotypes, are highly assisted in the forensic sciences to facilitate the identification process. All these techniques require the tissues and fluids to be in a relatively optimal state of preservation that enables both the extraction and amplification of genetic material.73,74 Unfortunately, it is common for there to be extreme physical and environmental characteristics at the forensic investigation site that do not facilitate this preservation. In addition, the genetic material itself is highly susceptible to degradation, preserves poorly over prolonged periods of time and samples are commonly contaminated in forensic contexts, which can limit the use of these tools in some cases.73,75
As has already been observed, in contrast to DNA, proteins are molecules that are highly resistant to physical and environmental conditions, and preserve well over long periods of time.59 Likewise, in the sequence of diverse proteins genetic variations can be found between one individual and another in the form of single-amino acid polymorphisms, which give rise to genetically variable peptides that are the direct product of non-repeated single nucleotide polymorphisms that are present in exonic regions of DNA. Therefore, based on the proteomic analysis of these peptides with genetic variation, individuals in certain population groups can be biogeographically located, and it can even be a useful tool in individualising the identification process.76,77
Several studies have addressed the possibility of using these peptides with genetic variation as auxiliary tools for identifying single nucleotide polymorphisms in the absence of genetic material in the forensic context, using different matrices such as hair and bone tissue. After protein extraction and enzymatic digestion, the multiple peptides are analysed by high efficiency liquid chromatography coupled with mass spectrometry, and bioinformatic analysis is performed to identify possible peptides with genetic variation.77,78 These peptides must be classified and analysed in such a way as to rule out false positives caused by MPT and/or environmental false positive such as methionine or cysteine, deamination of asparagine (N) or glutamine (Q), and the hydroxylation of amino acids such as proline, among others. This bioinformatic analysis, together with a statistical analysis, demonstrates the occurrence of polymorphism events, which enables the phylogenetic distance between individuals to be calculated, allowing individuals to be assigned within specific biogeographic al groups.76,78 This, in practice, may facilitate the classification of suspects and/or victims into a given population group. In addition, the identification and characterisation of these peptides with genetic variation together with an analysis of the expression pattern in the protein profile could theoretically allow the identification process to be individualised. Although, in this regard, diverse factors, such as dietary habits, environmental factors and age, among others, can generate enough variations in the gene expression profile, so that individual identification would be difficult in some cases. Nevertheless, identifying these peptides with genetic variation is outlined as a powerful tool in the forensic field, which currently enables the identification of biogeographical groups and, in the near future, of individuals in cases where the quality and quantity of the genetic material is not optimal for the processes of extraction and analysis of DNA sequences.76–78
In addition to this analysis that can result in determining the biogeographical group of subjects and their possible individual identification, the use of proteomic tools, both in ancient and contemporary remains, can also enable sex to be determined. Recently, it has been demonstrated that the analysis of amelogenin peptides by means of high performance liquid chromatography coupled with mass spectrometry enables this protein's different isoforms to be identified. Since amelogenin is coded in genes that are found in the sex chromosomes, its different isoforms are specifically associated with each of the sex chromosomes, providing a novel tool that can facilitate determination of the sex of individuals under study, whose genetic material is not of optimal quality.79
Limitations and challengesProteomics has proved a powerful tool in detecting molecules that have potential as biomarkers in the forensic field.30,76 Like any methodology, it presents a series of limitations and technical difficulties.80
Although several challenges remain, proteomics is emerging as a tool that, together with existing technologies, can enhance the acquisition and analysis of biological information with a high value from a medico-legal perspective.10 From the forensic perspective, we consider that there are two limiting factors to take into account: the first, referring the proteome itself to be analyzed and the second, referring to the limitations of the analytical methods used.
One of the main difficulties to consider is the nature of the sample, since in many forensic scenarios samples may be scarce and poorly preserved. This implies that protein extraction and isolation strategies are required that reduce contamination and degradation, while reducing the formation of unwanted MPT.14,81 In addition, the scenario from which the samples were recovered must also be considered, as environmental conditions can modify the preservation of proteins present in different types of tissue.58
Furthermore, it is important to bear in mind that proteomic analysis of samples in the forensic context is relatively recent. Therefore interindividual or intraindividual variation can affect the results obtained.30 This means that strict protocols must be developed for the use of these new strategies regarding the nature of the sample, the anatomical origin of the sample and the transport and preservation conditions. It is also important to advance in characterising new highly reproducible protein markers that will allow possible variations from one individual to another to be corrected.10,30
In some contexts, such as the analysis of archaeological or skeletal remains, it is possible that few proteins are recovered, which implies that a superficial analysis of the data could result in the loss of forensically valuable biological information.10,59 Therefore, proteomic analysis in the forensic area must be undertaken by experts using robust and specialised analytical platforms, which entails costs both in the training of specialist personnel and in the acquisition of cutting-edge technologies.
ConclusionsAlthough the interaction between proteomics and forensic sciences is relatively recent, there has been an increase in the number of publications and applications that this group of strategies could have in the medico-legal field. As we have seen, proteomics can address a host of questions that are crucial in understanding the forensic scene in a highly reproducible manner, with high quality standards, while reducing the possibility of false identification due to the contamination or degradation of samples.18,22,59 Employing these strategies in forensic laboratories could broaden the scope in the acquisition and analysis of biological data, which would enable not only the management of large volumes of information, but also the detailed interpretation of this information in different contexts, following a highly efficient and reproducible workflow, providing highly precise quantitative evidence, which forensic experts can interpret thanks to the wide variety of bioinformatic tools available.7,10,57
Possible applications in the forensic context range from the identification of samples found at a crime scene, to the precise diagnosis of diseases, metabolic or toxicological conditions of individuals related to the forensic scene, and even the determination of the biogeographical group to which they belonged, from a large number of matrices of biological and non-biological origin such as blood, semen, bone, tissue, hair, fibres or substrates such as soil or sand. The results provided by proteomics are fast and highly reliable because they rely on robust databases and statistical analyses that enable the reliable interpretation of information and with low likelihood of error.3,35,56,82
Finally, although many challenges remain, the use of these new technologies for identifying and quantifying protein markers, combined with the use of DNA and RNA markers that are already known, as well as the development of mass spectrometry technologies for proteins, can be projected into a new stage of forensic sciences, where the biological information provided by physical evidence enables us to establish not only the identity of the subjects, but also quantitative information on various events such as the health status of the individuals, their nutritional patterns, environmental conditions, migration events and movement of individuals involved in a forensic scene.
FinancingThis paper was made possible thanks to the Postdoctoral Grant awarded by the General Directorate of Academic Personnel Affairs of the UNAM.
Conflict of interestsThe authors have no conflict of interests to declare.
DMRD would like to acknowledge the support received from the Departments of Amphitheatre and Microbiology and Parasitology of the Faculty of Medicine of the National Autonomous University of Mexico (UNAM).
Please cite this article as: Díaz Martín RD, Camacho-Martínez Z, Ambrosio Hernández JR, Valencia-Caballero L. La proteómica como una nueva herramienta en las ciencias forenses. Rev Esp Med Legal. 2019;45:114–122.