Cerebral cavernous malformations (CCMs; OMIM 116860) are enlarged vascular cavities without intervening brain parenchyma whose estimated prevalence in the general population is between 0.1% and 0.5%. Familial CCM is an autosomal dominant disease with incomplete clinical and radiological penetrance. Three genes have been linked to development of the lesions: CCM1/KRIT1, CCM2/MGC4607, and CCM3/PDCD10.
DevelopmentThe aetiological mutation is not detected in a large percentage of cases and new approaches are therefore needed. The aim of this review is to analyse current molecular techniques and the possible mutations or variations which can be detected in a molecular genetics or molecular biology laboratory. Likewise, we will analyse other alternatives that may help detect mutations in those patients showing negative results.
ConclusionsA molecular diagnosis of CCMs should provide at least the copy number variation and sequencing of CCM genes. In addition, appropriate genetic counselling is a crucial source of information and support for patients and their relatives.
Las malformaciones cavernosas cerebrales (CCM; OMIM 116860) son engrosamientos cavernosos vasculares sin intervención del parénquima cerebral con una prevalencia estimada en la población general del 0,1-0,5%. La cavernomatosis cerebral presenta un patrón de herencia autosómico dominante con penetrancia clínica y radiológica incompleta. Tres genes se han asociado al desarrollo de lesiones: CCM1/KRIT1, CCM2/MGC4607 y CCM3/PDCD10.
DesarrolloLa mutación responsable no es detectada en un alto porcentaje de casos, por lo que nuevos enfoques son necesarios para su detección. En esta revisión se analizan las técnicas actualmente utilizadas y las posibles mutaciones o variantes que pueden ser detectadas en un laboratorio de genética molecular o biología molecular. Asimismo, se analizan alternativas que pueden ser abarcadas para la detección de mutaciones en aquellos pacientes en los que los estudios hayan resultado negativos.
ConclusionesEl diagnóstico molecular de la cavernomatosis cerebral debe incluir al menos la detección del número de copias y la secuenciación de los genes CCM. Finalmente, ofrecer un adecuado consejo genético es crucial para proporcionar información y apoyo a los pacientes y familias que padecen la enfermedad.
Cerebral vascular malformations encompass a wide range of relatively frequent lesions with very different natural history and clinical significance. The effects of lesions range from completely benign conditions to severe incapacitating neurological disorders or even death in a significant percentage of patients. The most common lesions are arteriovenous malformations and cerebral cavernous malformations (CCMs; OMIM 116860), which affect the central nervous system almost exclusively. Symptoms associated with these vascular malformations include epilepsy, focal neurological signs, and haemorrhagic infarcts due to rupture of the lesion.
CCMs are venous and cannot be detected using angiography, which explains why they are referred to as angiographically silent. Multiple terms have been used to designate these lesions, including cavernous angioma, cavernoma, cavernous haemangioma, and cavernous venous malformation. Their prevalence is estimated at 0.1% to 0.5%.1,2 CCMs are caused by enlargement of a well-defined collection of blood vessels and have no parenchymal involvement.3
Lesions may be sporadic or hereditary. Familial CCM has an autosomal dominant inheritance pattern with incomplete clinical and radiological penetrance. Some cavernomas are clinically silent whereas others may even cause seizures, haemorrhages, or focal neurological signs. CCMs account for 5% to 15% of all vascular malformations of the central nervous system. Cavernomas may be asymptomatic; in these cases, they may be detected by chance during an MRI scan. However, 50% to 70% of all cavernomas manifest clinically between the second and the fourth decade of life, regardless of sex. CCMs have also been described in paediatric patients.4 The actual incidence of CCMs is difficult to estimate, given that these malformations may be mistaken for other types of vascular malformations.3 At present, we only have estimated data of the general population drawn from autopsy and MRI studies. Two studies carried out in 1991 estimated prevalence of CCMs at 0.39% and 0.47%, based on a large series of MRI scans.5,6 Later studies have confirmed these data; prevalence of CCMs is currently estimated at 0.1% to 0.5% of the general population.1,2
CCMs may cause local neurological symptoms, such as epileptic seizures, or general symptoms, including headache and haemorrhagic cerebrovascular accidents. Intracranial bleeding is the most feared complication of CCMs, as it may result in severe disability or even death.7 CCMs are currently detected using T2-weighted gradient-echo MRI sequences, given that they may present with similar clinical symptoms to those of other vascular malformations or cerebral microhaemorrhages of different aetiology. Patients with CCMs may be classified in 2 groups:
- 1.
Familial CCMs. More than one patient in the family or only one patient with multiple lesions (multiple CCMs).
- 2.
Sporadic CCMs. A single patient in a family with only one cavernoma on gradient-echo MRI sequences.
Some studies have suggested that 75% of cases of sporadic CCMs are in fact familial and display asymptomatic lesions, masking an autosomal dominant pattern.8
DevelopmentGenetic diagnosisAt present, mutations are not identified in all patients with CCMs; they are detected in 78%9 to 40%10,11 of cases, according to recent studies. As a result, CCM molecular screening cannot be used to rule out the disease. Differences in these detection rates may be explained by the inclusion of patients with a history of cerebral haemorrhage of unknown aetiology. The Spanish Society of Neurology recommends conducting genetic testing of CCM genes in patients diagnosed with stroke.12
Three genes have been associated with CCMs: CCM1/KRIT1, located on the long arm of chromosome 7 (7q21–q22)13,14; CCM2/MGC4607, on the short arm of chromosome 7 (7p13)15; and CCM3/PDCD10, on chromosome 3 (3q26.1).16 Nearly 200 mutations have been described in patients with CCMs and a low risk of recurrence. In Spain and Portugal, however, a 14 bp deletion in exon 5 of CCM2 was found to be highly prevalent.17 In contrast, the mutation c.1363T>C in CCM1, which is highly prevalent in the Hispanic population,14 was not detected in Spanish patients.18 In the Spanish population, 56% of all mutations found in familial cases of CCMs affected CCM1, with 33% and 6% affecting CCM2 and CCM3, respectively.10 The mutation responsible for CCMs is yet to be detected in a high percentage of families. Large deletions have been detected in all 3 CCM genes in patients with CCMs.11,19–21
These 3 genes code for scaffold proteins involved in different processes, including angiogenesis and vasculogenesis. These 3 proteins interact to form a ternary complex. Mutations or variants leading to protein function loss are therefore considered pathogenic. The pathogenicity of such mutations is not easy to determine, however. Patients with CCMs may carry the following mutations: nonsense mutations, frameshift mutations, splice site mutations, missense mutations, silent mutations, and large deletions.
Nonsense and frameshift mutationsNonsense mutations introduce a premature stop codon, for example c.902C>G (p.S301X) in CCM1, which results in a truncated protein of 301 amino acids rather than 736. Frameshift mutations are caused by insertions, deletions, or indels (insertions and deletions in the same region) and may result in a premature stop codon. In CCM1, for example, insertion c.968_971dupCACC (p.Ile325ThrfsX11) results in a truncated protein of 336 amino acids. This is the most frequent type of pathogenic mutation.10,11
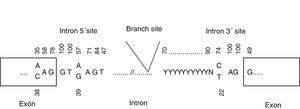
Splice site mutationsThere are 2 types of splice site mutations: (a) those affecting the invariant sequence (intron bases 1 and 2, adjacent to the splice site) and (b) those affecting the consensus sequence (exon bases 1-3 and intron bases 3-8 from the splice site) (Fig. 1). Mutations in the invariant sequence always alter splicing, which may lead to partial or complete exon skipping or to partial or complete intron retention in the mature transcript. This, in turn, results in a truncated or shorter protein, depending on whether the reading frame is affected. This explains why splice site mutations are considered pathogenic. In these cases, cDNA analysis is recommended to determine the exact consequences of the mutation.
On the other hand, mutations in the consensus sequence may or may not be pathogenic, and are therefore considered variants of uncertain significance. A cDNA study is essential to determine whether they are pathogenic. This type of study identifies the activation of a cryptic splice site, that is, a splice site located in a part of the transcript other than the wild-type splice site, resulting in a truncated protein. Cryptic splice site activation may be assessed in silico with the Human Splicing Finder (http://www.umd.be/HSF/).22
Missense and silent mutationsMissense mutations are caused by a change in one amino acid. In the case of silent mutations, protein-level alterations do not take place. Both mutations should be regarded as variants of uncertain significance; cDNA analysis is therefore recommended to assess cryptic splice site activation. Several studies into CCMs have found missense mutations activating cryptic splice sites,10,23–25 as well as silent mutations.15,26,27 The pathogenicity of these mutations may also be explained by an aberrant mutated protein subject to degradation; in this case, the normal allele would only be observed in cDNA.26 When both alleles are observed in cDNA, silent mutations are considered non-pathogenic.10,11 This is not the case with missense mutations, however: these would still be considered to be of an uncertain significance. Several bioinformatic tools, such as PolyPhen-2 and SIFT, can be used to predict the effect of a mutation on protein function using data on protein structure and sequence.28,29 However, proteomic studies are necessary to confirm the pathogenicity of a given mutation.
Large deletionsThe introduction of multiplex ligation-dependent probe amplification (MLPA) (P130 and P131, MRC-Holland, the Netherlands), which permits simultaneous PCR amplification using only one primer pair, has enabled the detection of deletions of more than one exon, a not infrequent occurrence. Quantitative multiplex PCR of short fluorescent fragments has also been used recently to detect deletions.11 Almost 40 deletions have been detected to date in CCM genes.10,11,16,20,21,27,30 These deletions cannot be detected using direct sequencing analysis and should be considered pathogenic, given that they tend to result in incomplete transcription.
Polymorphisms and intronic variantsSeveral polymorphisms may be identified during sequencing; these may appear both in exons (missense or silent mutations) and in introns. The first course of action in the case of these variants should be to consult a database such as the 1000 Genomes Project database (http://browser.1000genomes.org/index.html). Some of these polymorphisms have been associated with increased risk of CCMs, whereas others have been found to predispose patients to potentially disabling symptoms (headache) rather than to conditions that are potentially harmful to health (epilepsy, brain haemorrhages).24
A cDNA analysis should be conducted in patients with non-polymorphic intronic variants located outside splice sites in order to assess cryptic splice site activation. As reported in the literature, variants c.1564-14C>G and c.2026-12A>G in CCM1 cause skipping of exons 15 and 18, respectively, resulting in a truncated protein in the first case (p.I522X) and a shortened protein in the second (p.L677_A715del).11
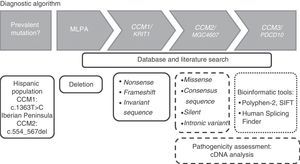
Genotyping strategyThe genotyping strategy may vary depending on the patient's ethnicity. As mentioned previously, the mutation c.1363T>C in CCM1 may be detected in Hispanic populations whereas a 14 bp deletion in exon 5 of CCM2 (c.554_567del) may be detected in people from the Iberian Peninsula. As a general rule and based on our own experience, we propose following the algorithm shown in Fig. 2 for the molecular study of CCMs.
The first stage of the process is to search for deletions in the 3 CCM genes using MLPA kits (P130 and P131, MRC-Holland, the Netherlands). If the number of copies is normal, the adjacent exonic and intronic regions in the CCM genes should be analysed using Sanger and serial sequencing. Sequencing should begin with CCM1, followed by CCM2 and finally CCM3. The study is completed upon detection of variants introducing a premature stop codon (nonsense or frameshift mutations) or located in the invariant sequence of a splice site, given that these mutations cause loss of protein function and are therefore pathogenic. Sequencing should not be stopped on detection of a missense mutation, a silent mutation, or an intronic variant, given the uncertain significance of these mutations. In these cases and in splice site mutations, cDNA analysis is strongly recommended for the assessment of cryptic splice site activation.
Alternatives and future perspectivesIn view of the fact that mutations responsible for CCMs go undetected in a high percentage of patients, other forms of genetic diagnosis should be considered.
- (i)
cDNA analysis of the 3 CCM genes. Using this alternative, Riant et al.31 detected a 99-bp insertion in the nucleic acid sequence of the complete transcript of CCM1 corresponding to intron 2. Subsequent DNA analysis detected a deletion (c.262+132_262+133delAA) causing the activation of an adjacent splice site. The inclusion of this fragment modifies the reading frame, introducing a stop codon which results in a truncated protein. We should highlight that this mutation would not have been detected with the algorithm presented in Fig. 2.
- (ii)
As a general rule, 5′ and 3′ UTRs are not studied. These regions have a considerable influence on mRNA stability.32,33 The 5′ UTR in CCM1 is especially complex, as it can differentially transcribe up to 30 variants with different combinations of non-coding exons. A recent study described 2 contiguous mutations in the 3′ UTR of CCM1 in the same patient, which may have caused interaction with miRNA24 and consequently decreased KRIT1 expression. Mutations in these regions may therefore alter mRNA stability and decrease the production of functional protein. However, confirming the pathogenicity of a mutation is not straightforward, given the difficulty of obtaining endothelial cells from the lesions of patients with CCMs.
- (iii)
At present, the new massively parallel sequencing techniques (next generation sequencing, NGS) are increasingly common in the private sector but are yet to be routinely used in public laboratories. These novel techniques increase the performance and speed of data generation and dramatically reduce the costs associated with base-by-base sequencing. Furthermore, they enable the 3 CCM genes to be analysed simultaneously. Analysing DNA and cDNA in the same run represents an interesting alternative. No data have been published to date on CCM mutation detection using these techniques. Therefore, each laboratory should perform a cost-benefit analysis for the technology included in their algorithm. High-resolution melting analysis34 may reduce overall cost by sequencing only those fragments with changes in the melting curve.
- (iv)
It has been suggested in recent years that a new gene may be involved in the pathogenesis of CCMs.35 Whole-exome sequencing using NGS may be useful in patients with a family history of CCMs and negative genetic testing results. Given their uncertain significance, the mutations identified with this technique should be validated clinically. A detailed description of patients’ clinical and radiological features is therefore essential.
Genetic counselling by a specialist should provide patients and/or their families with information on the risks of CCMs, possible treatments, and the risk of hereditary transmission of the disease. More specifically, genetic counselling should consider the following: (1) identifying the mutation; (2) determining whether the germline mutation is familiar or sporadic; (3) analysing clinical penetrance and determining, with the help of the physician, whether the patient presents a single malformation or multiple CCMs; (4) offering a molecular study to first-degree relatives (the study should be requested by patients after being informed about the risks of CCMs); (5) studying the proband's parents first (this is particularly relevant in the case of children at risk; in these cases, genetic counsellors should be even more cautious); (6) particular considerations must be taken when providing genetic counselling, since the lesion may be surgically resected and symptoms treated; and (7) genetic counselling with molecular diagnosis should not be provided to pregnant women when the mutation has not been identified.
ConclusionsMolecular diagnosis of CCMs should at least provide the number of copies and sequencing of CCM genes. Other alternatives may be considered when no mutation causing the disease is found. In a large number of patients, mutations are not detected; a molecular study cannot therefore rule out the disorder. Genetic counselling is an essential source of information and support for patients and their families.
Conflicts of interestThe authors have no conflicts of interest to declare.
Please cite this article as: Mondejar R, Lucas M. Diagnóstico molecular de cavernomatosis cerebral. Neurología. 2017;32:540–545.