Seroclearance of hepatitis B e antigen (HBeAg) is an important treatment goal for patients with chronic hepatitis B (CHB). This study developed a nomogram for predicting HBeAg seroclearance in CHB patients treated with nucleos(t)ide analogues (NAs).
Patients and MethodsFive hundred and sixty-nine CHB patients treated with NAs from two institutions between July 2016 to November 2021 were retrospectively included. One institution served as the training set (n = 374) and the other as the external validation set (n = 195). A predictive nomogram was established based on cox regression analysis.
ResultsThe overall HBeAg seroclearance rates were 27.3 and 21.5 % after the median follow-up of 100.2 weeks and 65.1 weeks in the training set and validation set, respectively. In the training set, baseline aspartate aminotransferase, gamma-glutamyl transpeptidase, HBeAg, and hepatitis B core antibody levels were independently associated with HBeAg seroclearance and were used to establish the HBEAg SeroClearance (ESC)-nomogram. The calibration curve revealed that the ESC-nomogram had a good agreement with actual observation. The ESC-nomogram showed relatively high accuracy for predicting 48 weeks, 96 weeks, and 144 weeks of HBeAg seroclearance in the training set (AUCs: 0.782, 0.734 and 0.671) and validation set (AUCs: 0.699, 0.718 and 0.689). The patients with high ESC-nomogram scores (≥ 79.51) had significantly higher cumulative incidence of HBeAg seroclearance and seroconversion than patients with low scores (< 79.51) in both sets (P < 0.01).
ConclusionsThe novel ESC-nomogram showed good performance for predicting antiviral efficacy in HBeAg-positive CHB patients with NAs treatment.
Chronic hepatitis B virus (HBV) infection remains a serious global health problem and is a major cause of liver fibrosis, cirrhosis, and hepatocellular carcinoma (HCC) [1,2]. Functional cure characterized by hepatitis B surface antigen (HBsAg) seroclearance with or without antibody against HBsAg (anti-HBs) positivity is considered an optimal endpoint of antiviral treatment for patients with CHB [3,4]. However, it is reported that HBsAg seroclearance almost exclusively occurs in hepatitis B e antigen (HBeAg)-positive CHB patients treated with nucleos(t)ide analogues (NAs), with 8-year HBsAg loss rate of 0.13 % [5]. Alternatively, HBeAg seroclearance is a satisfactory endpoint for patients with HBeAg-positive CHB, which represents a partial immune control of chronic HBV infection [4]. Moreover, sustained HBeAg seroclearance is correlated with an improved prognosis including sustained virus inhibition, a higher probability of HBsAg seroclearance and seroconversion, which is considered a permanent clinical remission of CHB [6–10].
NAs are the most commonly used antiviral drugs and have a potent efficacy in inhibiting HBV replication for patients with CHB [10]. Nevertheless, the efficacy of NAs is limited for HBeAg seroclearance. Previous studies reported that only 10%-20% of HBeAg-positive CHB patients achieved HBeAg seroclearance after 1-year treatment with NAs [11–14]. Hence, there is an urgent need to identify serum biomarkers that can accurately predict the HBeAg seroclearance. Previous studies have found that several biomarkers including baseline HBV DNA, alanine aminotransferase (ALT), HBsAg, HBeAg, platelet (PLT), gamma-glutamyl transpeptidase (GGT) as well as anti-hepatitis B core antibody (HBcAb) levels were associated with HBeAg seroclearance [15–21]. Although some predictive models have been established for assessing HBeAg seroclearance, most of them had a limited sample size or concentrate on the response to interferon [22–24]. There are lacking developing multi-parameter scoring models to better predict HBeAg seroclearance for NAs treatment. In this study, we aimed to develop and externally validate a novel nomogram for predicting the probability of HBeAg seroclearance in patients with CHB treated with NAs.
2Material and Methods2.1Study populationFive hundred and sixty-nine treatment-naïve HBeAg-positive CHB patients who received first-line NAs (entecavir [ETV], tenofovir disoproxil fumarate [TDF], and tenofovir alafenamide fumarate [TAF]) treatment were retrospectively included from Nanjing Drum Tower Hospital (Nanjing, China) and Huai'an No. 4 People's Hospital (Huai'an, China) between July 2016 to November 2021. Patients concurrent with the following diseases were excluded from this study: other viral hepatitis, including hepatitis A, hepatitis C, hepatitis D, hepatitis E; acquired immunodeficiency syndrome, primary biliary cirrhosis, autoimmune hepatitis, alcoholic hepatitis, nonalcoholic fatty hepatitis and metabolic liver diseases, HCC and liver failure. The Nanjing cohort serves as the training set and Huai'an cohort as the external validation set.
2.2Data collection and definitionBaseline laboratory parameters were collected from electronic medical record system, including blood routine examination, liver function tests, and serological markers of HBV. The ARCHITECT assay (Abbott Gmbh, Wiesbaden, Germany) was employed to detect the quantitative HBV serological markers. A positive HBsAg result was defined as a value exceeding 0.05 IU/ml. Additionally, if the HBsAg level was greater than 250 IU/ml, further testing was conducted using a stepwise dilution ranging from 1:20 to 1:1,000. A positive HBeAg result was determined by a sample/cutoff (S/CO) ratio of ≥ 1.0, while a positive HBeAb result was indicated by an S/CO ratio lower than 1.0. The detection of HBV DNA was carried out using either the Cobas TaqMan HBV test (Roche, Basel, Switzerland) or the TaqMan polymerase chain reaction assay (Daan Gene, Guangzhou, China). The lower limit of normal for HBV DNA detection was set at 20 IU/ml or 500 IU/ml, depending on the specific assay utilized. In addition, the diagnosis of cirrhosis was made by imaging tools, including ultrasound, computed tomography, or magnetic resonance imaging.
The liver fibrosis indexes used in this study were as follows: aspartate transaminase (AST) to PLT ratio index (APRI): (AST (U/L)/ULN of AST)/PLT count (109/L) × 100 [25]; the fibrosis-4 score (FIB-4): (age (years) × AST (U/L))/((PLT count (109/L) × (ALT (U/L))1/2) [26].
2.3Follow-up of patientsThe follow-up period of patients was 6 months or shorter interval during antiviral therapy. The primary endpoint was HBeAg seroclearance, which was defined as HBeAg loss regardless of anti-HBe status. The second endpoint was HBeAg seroconversion, which was defined as HBeAg loss and presence of anti-HBe. The follow-up time were calculated from the date of initiating antiviral therapy to the data of HBeAg loss and HBeAg seroconversion for patients with achieved HBeAg seroclearance and HBeAg seroconversion. The duration of follow-up for patients without HBeAg loss was calculated from the date of initiating antiviral therapy to the last date of follow-up.
2.4Statistical analysisContinuous variables were expressed by medians (interquartile range [IQR]), and categorical variables were expressed as the counts and percentages. The independent sample T test and Mann Whitney U test was used for comparing variables with and without normal distribution, respectively, and the comparison of categorical variables was conducted by Chi-square or Fisher exact test. Independent predictive factors of HBeAg seroclearance were identified by multivariate Cox proportional hazards regression analysis. A HBEAg SeroClearance (ESC)-nomogram for predicting HBeAg seroclearance based on these predictive factors was established based on training set. The predicting value of the ESC-nomogram was evaluated using the area under the receiver operating characteristic curve (AUC) and the calibration curve. Finally, according to the value of nomogram, all patients were divided into the high-score and low-score groups according to “surv_cutpoint” function from “survminer” package, and the Kaplan-Meier method with a log-rank test was used to estimate the cumulative HBeAg seroclearance and seroconversion. Pvalue < 0.05 was considered as statistical significance. Statistical analysis was conducted using Statistical Package for the Social Sciences version 24.0 software program (IBM, Armonk, NY, USA) and R software (version 4.2.0; R Foundation, Vienna, Austria; www.R-project.Org).
2.5Ethical statementThe ethics committees of Nanjing Drum Tower Hospital approved this study (approval No: 2008022) and the protocol was conducted following the Declaration of Helsinki guidelines. Since it was a retrospective design, the ethics committees granted a waiver of informed consent.
3Results3.1The characteristics of the study populationA total of five hundred and sixty-nine treatment-naïve patients with HBeAg-positive CHB were included in this study (374 patients from Nanjing cohort [training set] and 195 patients from Huai'an cohort [validation set]). The clinical features of patients were shown in Table 1. Overall, the median age of patients was 36.0 (IQR, 31.0–45.0) years, and 392 (68.9 %) patients were male. The median level of ALT, HBV DNA, HBsAg, and HBeAg were 108.0 U/L, 7.2 log10 IU/mL, 4.0 log10 IU/mL, and 3.0 log10 S/CO, respectively. More than half of patients were treated with ETV (62.4 %), followed by TDF (33.4 %) and TAF (4.2 %). During a median follow-up of 90.6 weeks, a total of 144 (25.3 %) patients achieved HBeAg seroclearance and 103 (18.1 %) patients achieved HBeAg seroconversion.
Baseline characteristics of the study population.
ALT, alanine transaminase; AST, aspartate transaminase; GGT, Gamma-glutamyl transferase; HBeAg, hepatitis B e antibody; HBV, hepatitis B virus; HBsAg, hepatitis B surface antigen; HBcAb, Hepatitis B Core Antibody; ETV, entecavir; TDF, tenofovir disoproxil fumarate; TAF, tenofovir alafenamide fumarate.
In training set, 102 (27.3 %) patients achieved HBeAg seroclearance after a median follow-up of 100.2 weeks. The age, gender, presence of cirrhosis and antiviral drugs were comparable between patients with and without HBeAg seroclearance. However, patients with HBeAg seroclearance had lower median levels of HBV DNA (6.9 log10 IU/ml vs. 7.5 log10 IU/ml, P < 0.001), HBeAg (2.8 log10 S/CO vs. 3.0 log10 S/CO, P = 0.001) and HBsAg (3.8 log10 IU/ml vs. 4.3 log10 IU/ml, P < 0.001) than non-seroclearance patients. Contrarily, patients with HBeAg seroclearance had significantly higher levels of ALT (159.5 U/L vs. 92.4 U/L, P < 0.001), AST (84.7 U/L vs. 51.9 U/L, P < 0.001), GGT (50.3 U/L vs. 33.0 U/L, P < 0.001), and HBcAb (9.6 S/CO vs. 8.9 S/CO, P < 0.001) than non-seroclearance patients. In terms of liver fibrosis index, patients who achieved HBeAg seroclearance showed higher APRI (1.2 vs. 0.8, P < 0.001) and FIB-4 (1.4 vs. 1.1, P = 0.008) values (Table 2).
Comparisons of baseline characteristics between patients with and without HBeAg seroclearance in training set.
ALT, alanine transaminase; AST, aspartate transaminase; GGT, Gamma-glutamyl transferase; HBeAg, hepatitis B e antibody; HBV, hepatitis B virus; HBsAg, hepatitis B surface antigen; HBcAb, Hepatitis B Core Antibody; APRI, aminotransferase to platelet ratio index; FIB-4, fibrosis-4 score; ETV, entecavir; TDF, tenofovir disoproxil fumarate; TAF, tenofovir alafenamide fumarate.
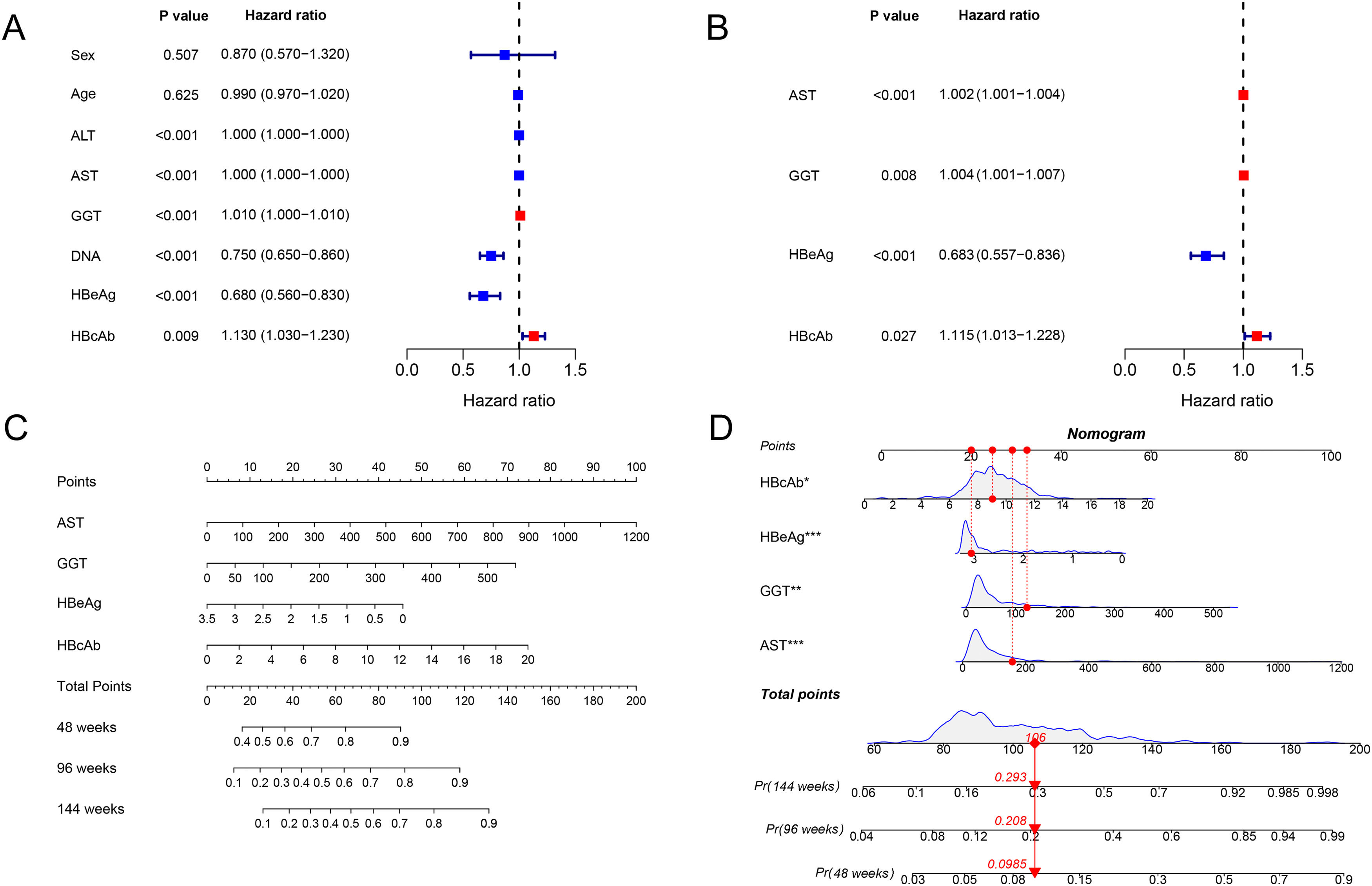
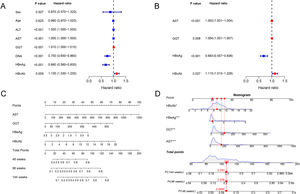
The associated factors of HBeAg seroclearance were identified by cox regression analysis in the training set. Univariate analysis revealed that ALT, AST, GGT, HBV DNA, HBeAg and HBcAb were associated with HBeAg seroclearance (Fig. 1A). These parameters were further included into multivariate analysis, which showed that AST (HR 1.002, 95 % confidence interval [CI] 1.001–1.004, P < 0.001), GGT (HR 1.004, 95% CI 1.001–1.007, P = 0.008), HBeAg (HR 0.683, 95 % CI 0.557–0.836, P < 0.001), and HBcAb (HR 1.115, 95 % CI 1.013–1.228, P = 0.027) were independent predictors of HBeAg seroclearance in HBeAg-positive CHB patients (Fig. 1B).
The forest plot demonstrated the univariate (A) and multivariate (B) analyses of variables for HBeAg seroclearance in HBeAg-positive chronic hepatitis B patients in training set. A nomogram for predicting 48, 96, and 144 weeks HBeAg seroclearance (C) and a dynamic nomogram depicting a patient as an example (D).
AST, GGT, HBeAg and HBcAb were used to develop a HBeAg seroclearance estimation ESC-nomogram (Fig. 1C). The point of each variable can be determined by drawing a line straight upward from each predictor to the point axis, and the total points can be calculated by summing single variable's point. The 48 weeks, 96 weeks, and 144 weeks HBeAg seroclearance can be found by drawing a line straight down from the total point axis. For example, a patient with the AST of 157.1 U/L, GGT of 123.1 U/L, HBeAg of 3.05 log10 S/CO, HBcAb of 9.04 S/CO, has 48 weeks, 96 weeks, and 144 weeks HBeAg seroclearance rate of 9.85, 20.8 and 29.3 %, respectively, according to this ESC-nomogram (Fig. 1D).
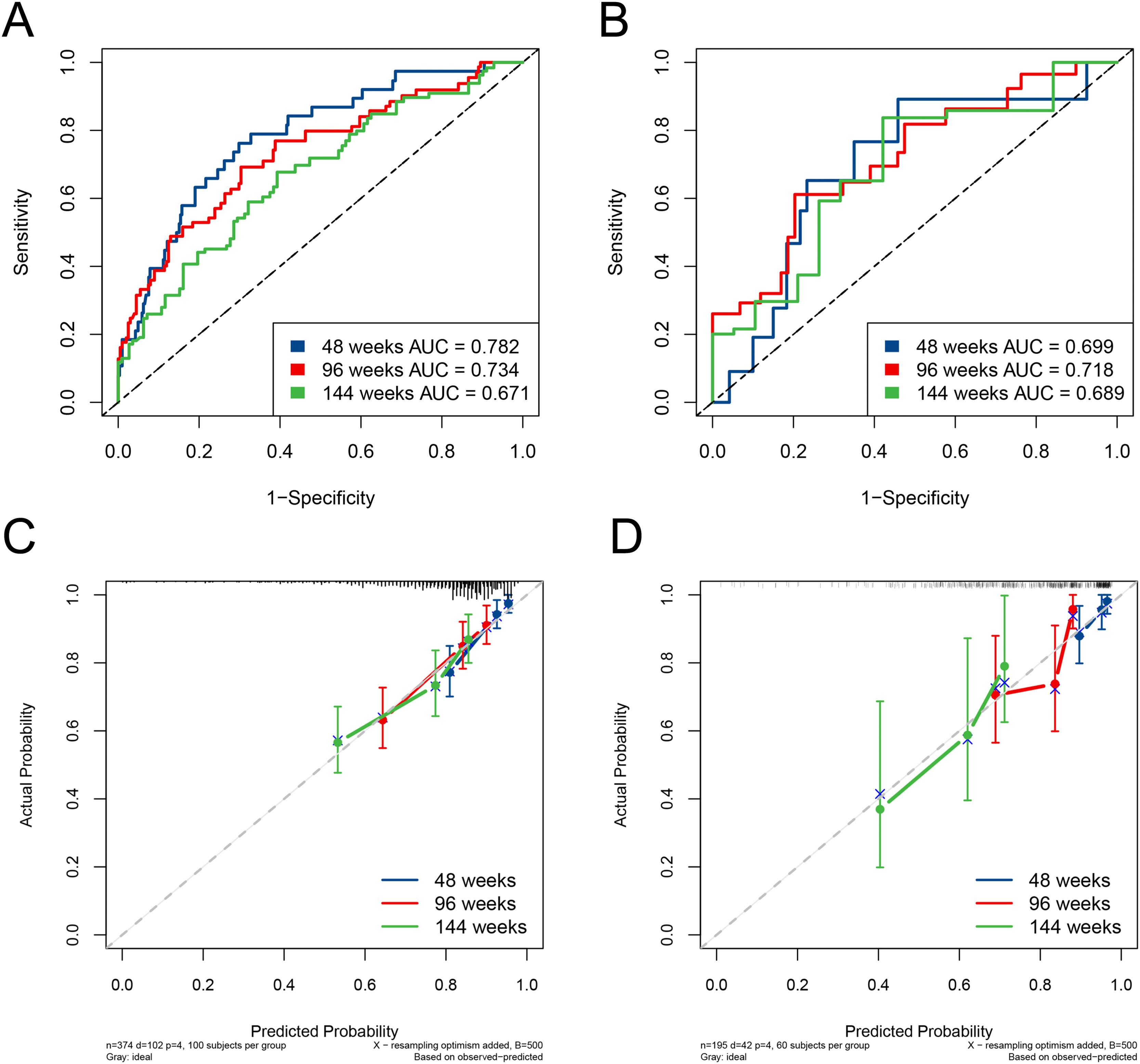
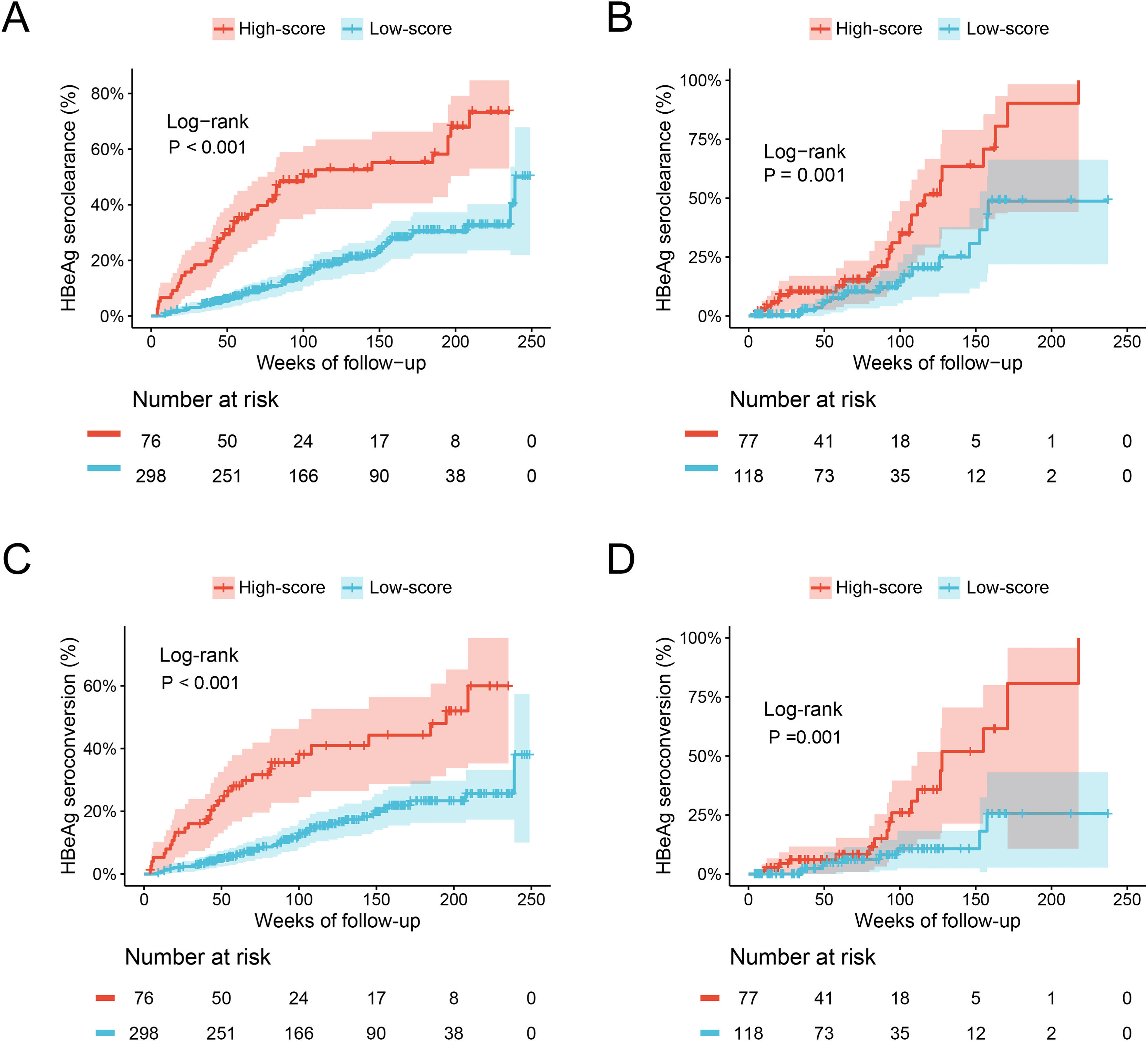
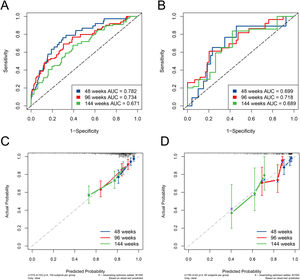
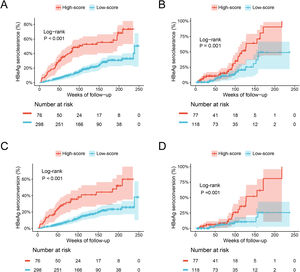
The ESC-nomogram showed a good accuracy in estimating HBeAg seroclearance in HBeAg-positive patients with CHB. The AUCs for predicting HBeAg seroclearance at 48 weeks, 96 weeks, and 144 weeks of treatment were 0.782 (95 % CI 0.706–0.858), 0.734 (95 % CI 0.661–0.808) and 0.671 (95 % CI 0.594–0.748) in training set, respectively (Fig. 2A). In addition, there were also good calibration curves for the HBeAg seroclearance estimation by the ESC-nomogram and the actual predicted probability on 48 weeks, 96 weeks, and 144 weeks after the antiviral therapy, respectively (Fig. 2C). The AUCs for the 48 weeks, 96 weeks, and 144 weeks HBeAg seroclearance were 0.699 (95 % CI 0.533–0.866), 0.718 (95 % CI 0.590–0.847) and 0.689 (95 % CI 0.535–0.842) in validation set, respectively (Fig. 2B). Similar well-calibrated results were observed in validation set (Fig. 2D). In addition, patients with high scores (≥ 79.51) were divided into high-score group, while patients with low scores (< 79.51) were divided into low-score group according “surv_cutpoint” function, which is an outcome-oriented methods providing a value of a cutpoint that correspond to the most significant relation with outcome (Figure S1). Kaplan-Meier analysis suggested that high-score group had significant higher cumulative HBeAg seroclearance and HBeAg seroconversion rates both in the training set (Fig. 3A and C) and validation set (Fig. 3B and D).
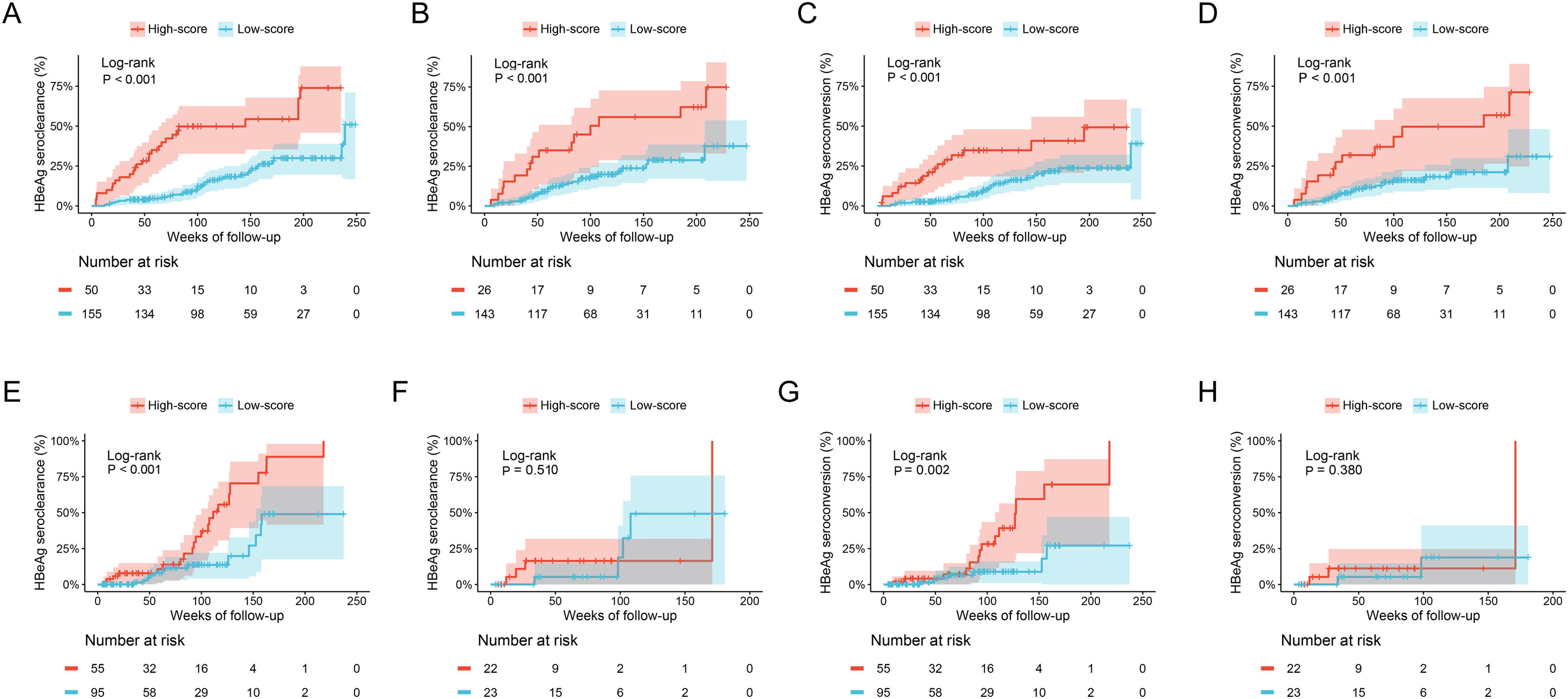
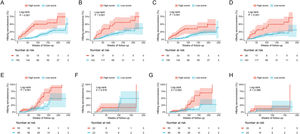
The cumulative HBeAg seroclearance and HBeAg seroconversion rates were also analyzed in patients treated with different drugs. Baseline characteristics of these patients between ETV group and TDF/TAF group were showed in Table S1. Patients in high-score group presented higher cumulative HBeAg seroclearance and HBeAg seroconversion rates than those of patients in low-score group in both ETV group (Fig. 4A and C) and TDF/TAF group (Fig. 4B and D) in training set. Similarly, in validation set, patients in high-score group had higher cumulative HBeAg seroclearance (Fig. 4E) and HBeAg seroconversion (Fig. 4G) rates than those of patients in low-score group in ETV group, while there was no significant difference in patients in TDF/TAF group (Fig. 4F and H).
Comparisons of cumulative HBeAg seroclearance and HBeAg seroconversion in different treatment groups between patients with high-score group and low-score group. Comparisons of cumulative HBeAg seroclearance in ETV group of training set (A) and validation set (E) and TDF/TAF group of training set (B) and validation set (F); Comparisons of cumulative HBeAg seroconversion in ETV group of training set (C) and validation set (G) and TDF/TAF group of training set (D) and validation set (H).
We further performed a subgroup analysis between patients with and without cirrhosis, as well as patients with and without ALT elevation. The comparison of clinical features was showed in Tables S2 and S3, respectively. The analysis suggested that the cumulative HBeAg seroclearance and HBeAg seroconversion rates in high-score group were significantly higher than low-score group in both patients without (Fig. S2A and S2C) and with cirrhosis (Fig. S2B and S2D) in training set. Similar results were observed in patients without cirrhosis (Fig. S2E and S2G) in validation set, while the cumulative HBeAg seroclearance and HBeAg seroconversion rates were comparable between high-score group and low-score group in patients with cirrhosis (Fig. S2F and S2H). In addition, patients in high-score group also had higher cumulative HBeAg seroclearance and HBeAg seroconversion rates than patients in low-score group in patients without (Fig. S3A and S3C) and with ALT elevation (Fig. S3B and S3D) in training set. In validation set, high-score group remains higher cumulative HBeAg seroclearance and HBeAg seroconversion rates than patients in low-score group in patients with ALT elevation (Fig. S3F and S3H), while there was no significant difference in patients without ALT elevation (Fig. S3E and S3G).
4DiscussionIn this study, we established a novel ESC-nomogram for predicting antiviral efficacy based on four routine parameters (AST, GGT, HBeAg, and HBcAb) in HBeAg-positive CHB patients treated with NAs. The ESC-nomogram could predict the possibility of HBeAg seroclearance and seroconversion after antiviral treatment of 48 weeks, 96 weeks, and 144 weeks with relatively high accuracy. The predicting performance was also validated in an external cohort.
HBeAg seroclearance has always been regarded as an ideal treatment endpoint for patients with CHB and suggests a transition of immune status of chronic HBV infection [4]. Numerous studies have reported that patients achieved HBeAg seroclearance early often have a better clinical outcome [27–29]. Accurately evaluating possibility of HBeAg seroclearance before antiviral treatment is necessary to formulate individualized treatment strategy for patients with CHB. However, there has been a lack of predictive method for HBeAg seroclearance in clinical practice. Several studies have developed some predicting models of HBeAg seroclearance. However, it is worth noting that these studies either exhibit restricted accuracy or lack external validation and subgroup analysis [30–32]. Therefore, our study developed and validated this ESC-nomogram based on routinely available clinical parameters with external validation which showed high accuracy in predicting HBeAg seroclearance.
Single parameters, including ALT, AST, GGT, HBeAg, HBcAb, and HBV DNA levels, have been identified to be associated with HBeAg seroclearance in patients with HBeAg-positive CHB after antiviral treatments [30,33–36]. However, in this study, several routine parameters were found to be associated with HBeAg seroclearance and included in this ESC-nomogram, including AST, GGT, HBeAg, and HBcAb. The AST is routinely measured in most patients with CHB and commonly associated with the severity of liver injury [37]. It has been confirmed that entecavir treatment successfully reduced the levels of ALT, AST, and HBV serological markers in previous study [38]. The present study revealed that the baseline AST level was an independent predictor of HBeAg seroclearance in CHB patients. It has been reported that elevated serum GGT levels were linked to bile duct damage, severe necroinflammatory activity, advanced fibrosis, or hepatic steatosis, which has emerged as a potential biomarker in the management of HBV infection [39–42]. In addition, it is also reported that increasing GGT levels have been identified to be an indicator of liver damage in CHB patients with hepatic steatosis [43]. Baseline HBeAg level was a well-known independent indicator of HBeAg seroclearance in CHB patients, and patients with lower HBeAg level had higher possibility of HBeAg seroclearance [22,44]. Numerous studies have reported that HBcAb level was an independent predictor of antiviral efficacy in patients with CHB [14,17,31,45]. Our previous study demonstrated that the HBcAb had good performance for predicting HBeAg seroconversion in CHB patients treated with NAs [14]. Serum HBcAb levels was reported to be significantly correlated with the severity of liver inflammation, which may be one possible mechanism of its association with antiviral efficacy [46,47].
Our results indicated that the ESC-nomogram was useful for assessment of the HBeAg seroclearance in CHB patients treated with NAs. The major advantages of this study included that this is a large cohort and the ESC-nomogram was validated by an external cohort. Nevertheless, the study also has several limitations. First, this is a retrospective study and selection bias might exist. Thus, prospective studies are needed to validate the performance of ESC-nomogram. Moreover, the predicting value of the ESC-nomogram for antiviral response of interferon needs to be validated in future studies. Third, the follow-up period is not long enough, especially for the validation cohort. Thus, the performance of the ESC-nomogram for long-term outcomes needs to be explored. In addition, HBV genotype data of patients were not available in this study. As previously reported, HBV genotype B or C are common in Asia CHB patients [48]. Thus, the predictive values of this nomogram in HBV-infected patients with different genotypes are not yet clear. Last, the sample size is relatively small, especially for TDF/TAF group. Thus, the ESC-nomogram should be validated in large cohort studies in the future.
5ConclusionsIn conclusion, this study established a novel ESC-nomogram in predicting HBeAg seroclearance in CHB patients treated with NAs. The application of the ESC-nomogram is used-friendly to identify patients with high HBeAg seroclearance possibility and guide the management of CHB.
Data availability statementThe data that support the study findings are available upon reasonable request from the corresponding authors (Chao Wu or Rui Huang).
FundingDr. Rui Huang wishes to acknowledge the supported from Nanjing Medical Science and Technique Development Foundation (Grant Nos. JQX21002 and QRX17121), Natural Science Foundation of Jiangsu Province (Grant No. BK20211004), and Clinical Trials from the Affiliated Drum Tower Hospital, Medical School of Nanjing University (2022-LCYJ-MS-07). Dr. Jian Wang wishes to acknowledge the support from the Clinical Trials from the Affiliated Drum Tower Hospital, Medical School of Nanjing University (2021-LCYJ-PY-43) and Nanjing Medical Science and Technique Development Foundation (Grant No. YKK21067). Dr. Juan Xia wishes to acknowledge the support from the Nanjing Medical Science and Technique Development Foundation (Grant No. YKK22073) and Clinical Trials from the Affiliated Drum Tower Hospital, Medical School of Nanjing University (2022-LCYJ-PY-49).
CRediT authorship contribution statementYan Gu: Formal analysis, Writing – original draft, Writing – review & editing, Data curation. Yao Zhang: Funding acquisition, Writing – review & editing, Data curation. Zhiyi Zhang: Funding acquisition, Writing – review & editing, Writing – original draft, Formal analysis, Data curation. Jian Wang: Methodology, Writing – review & editing, Writing – original draft. Qing Zhang: Funding acquisition, Writing – review & editing, Data curation. Shaoqiu Zhang: Funding acquisition, Writing – review & editing, Data curation. Yilin Liu: Funding acquisition, Writing – review & editing, Data curation. Jiacheng Liu: Funding acquisition, Writing – review & editing, Data curation. Juan Xia: Funding acquisition, Writing – review & editing, Data curation. Xiaomin Yan: Funding acquisition, Writing – review & editing, Data curation. Jie Li: Funding acquisition, Writing – review & editing, Data curation. Xingxiang Liu: Funding acquisition, Writing – review & editing, Conceptualization, Visualization, Data curation. Rui Huang: Methodology, Writing – review & editing, Conceptualization, Visualization. Chao Wu: Methodology, Writing – review & editing, Conceptualization, Visualization.