The impact of inactivated vaccines for COVID-19, BBIBP-CorV COVID-19, on the human immune system was not studied. This study investigates the immune response induced by the BBIBP-CorV COVID-19 vaccine.
MethodA total of 90 blood samples (5 ml each) were collected from participants (mean age: 20 years) in Basrah, Iraq. The study included 60 vaccinated individuals (38 males, 22 females), 60 days post-BBIBP-CorV vaccination and 30 unvaccinated controls (15 males, 15 females). Blood was divided: 2 ml in EDTA tubes for TLR7/TLR8 mRNA expression (RT-qPCR) and 3 ml in clotting activator tubes for serum isolation to measure IL-7, IL-10, IL-12, IL-18, and IL-21 levels (sandwich ELISA). Data were compared with controls and analysed for statistical differences. Volunteers with flu, COVID-19 symptoms, fever, chronic diseases, or immunocompromised conditions were excluded.
ResultsThe BBIBP-CorV vaccine did not disrupt cytokine production, with no significant differences in IL-7, IL-10, IL-12, IL-18, and IL-21 levels between vaccinated and unvaccinated groups after 60 days (p > 0.05). However, TLR7 and TLR8 mRNA expression was significantly upregulated (p < 0.05) in vaccinated individuals compared to controls, indicating enhanced innate immune activation without affecting cytokine balance. These findings highlight the vaccine's ability to stimulate immunity while maintaining normal cytokine levels.
ConclusionsThese results are particularly significant as they demonstrate that the BBIBP-CorV COVID-19 vaccine successfully induces immunological responses via TLR7 and TLR8 activation without abnormal effects on cytokine production. As a pilot study, these findings lay a strong foundation for future research.
No se ha estudiado el impacto de las vacunas no activadas contra la COVID-19, tal como BBIBP-CorV COVID-19, en el sistema inmunitario humano. Este estudio investiga la respuesta inmunitaria inducida por la vacuna BBIBP-CorV contra la COVID-19.
MétodoSe recogió un total de 90 muestras de sangre (de 5 ml cada una) de los participantes (edad media: 20 años) en Basrah, Irak. El estudio incluyó 60 individuos vacunados (38 varones, 22 mujeres) 60 días tras la vacunación con BBIBP-CorV, y 30 controles no vacunados (15 varones, 15 mujeres). Se dividió la sangre del modo siguiente: 2 ml en tubos EDTA para estudiar la expresión del ARNm de TLR7/TLR8 (RT-qPCR), y 3 ml en tubos activadores de coagulación para aislamiento sérico, a fin de medir los niveles de IL-7, IL-10, IL-12, IL-18 e IL-21 (ELISA tipo sándwich). Se compararon los datos con los controles, y se analizaron para encontrar las diferencias estadísticas. Se excluyeron los voluntarios con gripe, síntomas de COVID-19, fiebre, enfermedades crónicas o condiciones inmunocomprometidas.
ResultadosLa vacuna BBIBP-CorV no alteró la producción de citocinas, ni reflejó diferencias significativas de los niveles de IL-7, IL-10, IL-12, IL-18 e IL-21 entre los grupos de vacunados y no vacunados transcurridos 60 días (p > 0,05). Sin embargo, la expresión del ARNm de TLR7 y TLR8 mRNA estuvo significativamente aumentada (p < 0,05) en los individuos vacunados, en comparación con los controles, lo cual indica el aumento de la activación inmunitaria innata sin afectar al equilibrio de las citocinas. Dichos hallazgos destacan la capacidad de la vacuna para estimular la inmunidad, a la vez que mantiene los niveles de citocinas normales.
ConclusionesEstos resultados son particularmente significativos, ya que demuestran que la vacuna BBIBP-CorV contra la COVID-19 induce exitosamente las respuestas inmunológicas a través de la activación de TLR7 y TLR8, sin efectos anormales en la producción de citocinas. Como estudio piloto, estos hallazgos establecen un fundamento sólido para la investigación futura.
By late 2019, a novel coronavirus, severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2), emerged as the causative agent of coronavirus disease 2019 (COVID-19).1 The pandemic has had devastating effects on global health, society, and economies, necessitating urgent scientific and medical interventions. SARS-CoV-2 primarily causes pneumonia and upper/lower respiratory tract infections, with saliva immune barriers serving as the first line of defence.2 To overcome this pandemic, considerable effort was exerted to develop an effective vaccine, which is an immuno-biological substance designed to produce specific protection against SARS-CoV-2 antigens that evoke the immune response against viral infections.3 Protection stimulated by immunisation is mediated through a complex interplay between innate, humoral, and cell-mediated immunity.4 To quantify vaccine responses, measuring cytokine responses is one of the essential parameters alongside antibody titters, functional antibodies, antibody avidity, B and T cell activation, and lymphoproliferation.5 For instance, IL-7 is crucial for lymphocyte development and enhances vaccine-induced immune responses.6 IL-10 mitigates hyperinflammation by stimulating endogenous danger signals during tissue damage.7 IL-12, secreted by antigen-presenting cells (APCs), activates natural killer (NK) cells and promotes Th1 CD4+ cell differentiation, playing a vital role in antiviral immunity.8 IL-18, a pro-inflammatory cytokine, induces IFN-γ production and activates Th1, Th2, NK, and macrophage responses.9 IL-21, produced by CD4+ T-cells, particularly follicular helper T (Tfh) cells, regulates antibody production by B-cells and activates CD8+ T-cells.10,11 Notably, IL-21 responses correlate positively with SARS-CoV-2-specific B-cell responses and IgG antibody levels.12 Toll-like receptors (TLRs), part of the pattern recognition receptor (PRR) family, also play a pivotal role in detecting SARS-CoV-2 pathogen-associated molecular patterns (PAMPs). TLR7 and TLR8, localised on endosomal membranes, recognise single-stranded RNA (ssRNA) and trigger chemokine production to recruit immune cells, thereby controlling viral load.13,14 These mechanisms underscore the importance of understanding how vaccines influence immune components like interleukins and TLRs. Multiple vaccine platforms were developed, including whole-virus (live-attenuated or inactivated), viral vectors, nanoparticles, subunit proteins, RNA, DNA, and live-cell-based approaches.15 Despite widespread vaccination campaigns, the long-term immunity conferred by these vaccines remains under investigation. Few studies have explored the impact of COVID-19 vaccines on systemic and mucosal immunity.16–18 Measuring salivary biomarkers in vaccinated individuals could provide insights into mucosal immunity and its potential diagnostic applications.2 This study focuses on the BBIBP-CorV COVID-19 vaccine and its effects on IL-7, IL-10, IL-12, IL-18, and IL-21 cytokines, as well as TLR7 and TLR8 gene expression. To our knowledge, this is the first global study examining the BBIBP-CorV COVID-19 vaccine's impact on these immune markers, offering valuable insights into its efficacy and implications for individuals living with Long COVID. Further research is essential to better understand the vaccine's role in long-term immunity and health outcomes.
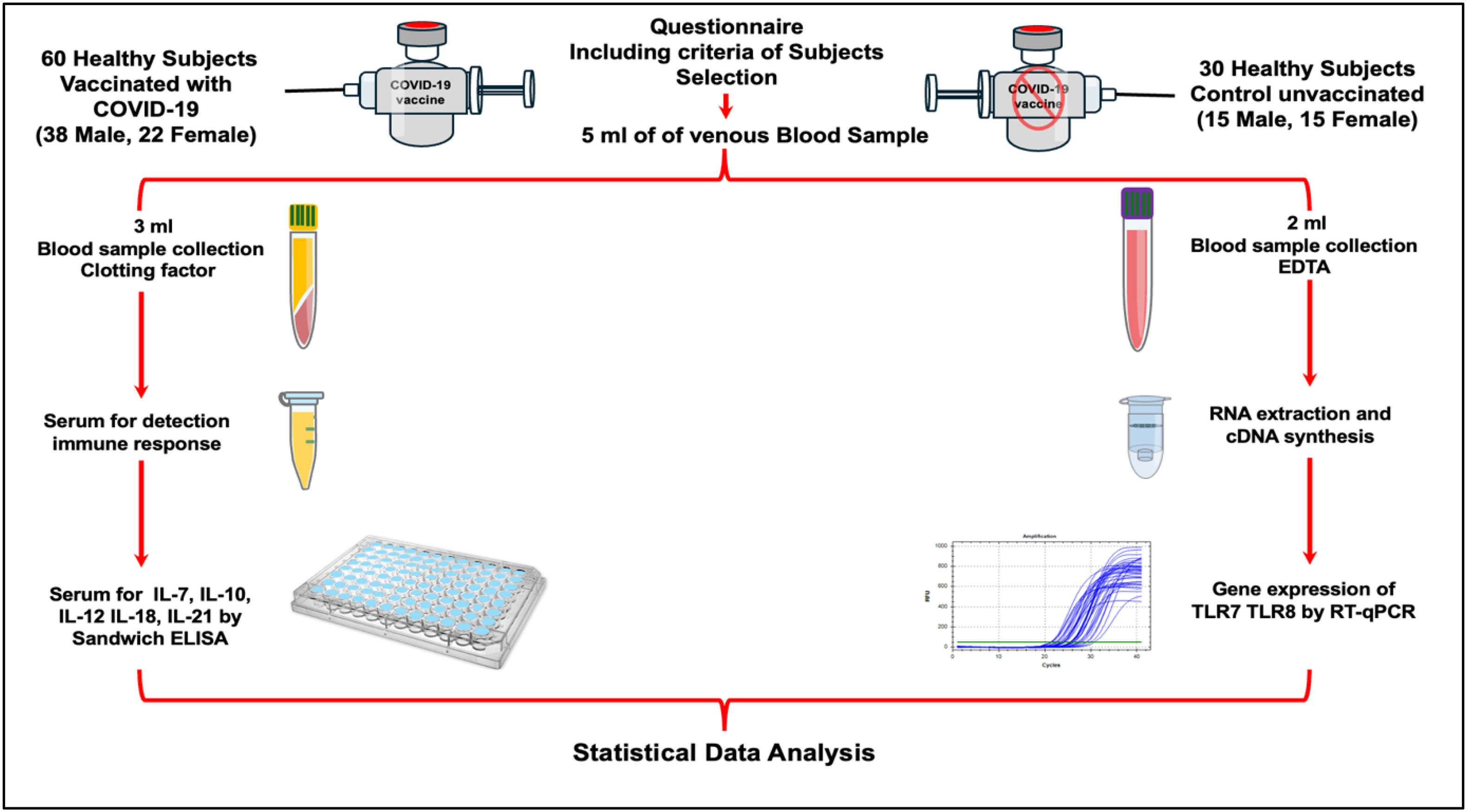
Materials and methodsStudy design and participation selectionThis study was conducted in approved COVID-19 vaccination centres in Basrah province, Iraq. Between April 2022 and October 2022, a total of 90 blood samples from two groups in this study were collected. Blood samples were collected 60 days after immunisation19,20 from the first group, which consisted of vaccinated participants who received the first dose of the BBIBP-CorV COVID-19 vaccine. This group included 60 participants (38 males and 22 females) who were enrolled in the study after providing informed consent for both the questionnaire and blood sample collection. The second group control group (Un-vaccinated) included 30 volunteers (15 Male, 15 Female) who had not received any of the COVID-19 vaccines. The study design, sample collections and testing methods were illustrated in Fig. 1.
Participant selection criteriaParticipant selection criteria participants were carefully screened to ensure the integrity of the study. Individuals exhibiting any symptoms of COVID-19, such as fever, cough, shortness of breath, or loss of taste or smell, were excluded from the study. Additionally, immunocompromised individuals, including those with conditions such as HIV/AIDS, cancer, or autoimmune diseases, were excluded to avoid confounding effects on immune responses. Participants undergoing immunosuppressive treatments, such as chemotherapy, long-term corticosteroid use, or biologic therapies, were also excluded, as these treatments could alter immune function and cytokine profiles. This rigorous selection process was implemented to ensure that the observed immune responses were attributable to the vaccine and not influenced by pre-existing conditions or treatments. Regarding Ethics Approval, the study was approved by the Scientific Committee of Biology Department, College of Science, University of Basrah. All participants provided informed consent to be included in the study.
Biological samplesA total of 5 ml of blood samples were collected from each participant in this study. The blood sample was divided into two tubes, the first, 2 ml placed in a labelled sterile EDTA tube to prevent blood clotting for RNA extraction. Immediately, EDTA tubes were gently inverted 10 times. Subsequently, 250 μl of whole blood sample was added to 750 μl of TransZol in a sterile clean 1.5-ml Eppendorf tube and vortexed well. Then, the samples were kept in deep freeze until use. The remaining 3 ml was placed in a clotting activator gel tube for serum isolation to measure cytokines level.
Cytokine analysisFor serum isolation, clotting activator gel tubes, 3 ml of whole blood, were left 15 min for clotting at room temperature and then centrifuged at 5000 rpm/10 min. The serum was collected and aliquoted into a sterile clean 1.5-ml Eppendorf tube 250 μl for each cytokine. Samples were measured by a duplicate for each sample using a Sandwich Enzyme-Linked Immunosorbent Assay (ELISA), following carefully the manufacturer's instructions. The concentration levels of five cytokines including IL-7 (Cat. No: E-EL-H0648), IL-10 (Cat. No: E-EL-H6154), IL-12 (Cat. No: E-EL-H0150), IL-18 (Cat. No: E-EL-H0253) and IL-21 (Cat No: E-EL-H2450) were measured.
RNA extractionUnder clean and sterile conditions, RNA was extracted from whole blood specimens using a TransZol up kit (Cat. No. ET111-01, TransGen biotech, China) following the manufacturer's protocol with modification. Briefly, 750 μl of TransZol up and 250 μl of whole blood specimens were vortexed immediately for 30 s. RNA was extracted using chloroform by mixing vigorously and then centrifuged at 10,000g for 15 min at 2–8 °C. As a result, the mixture separates into a lower organic pink phase, an interphase and an upper colourless aqueous phase, which contains the RNA. The upper colourless aqueous phase was carefully transferred to a clean Eppendorf tube, and then, around 200 μl of absolute ethanol was added. Next, the new mixture was transferred to a new spin column and washed with 600 μl of 70% ethanol. Total RNA concentration was measured using a NanoDrop Spectrophotometer (Avans Biotechnology, Taiwan).
RNA isolation, reverse transcription, and RT-qPCRTotal RNA was reverse-transcribed using reverse transcriptase and a cDNA synthesis GoScript™ Reverse Transcription System provided by (Promega company USA).
For evaluation of TLR7 and TLR8 gene expression, the RT-qPCR technique was conducted using the synthesised cDNA and Syber green chemistry (Cat. No. STD01B-M50h, SolGent, Korea). For gene amplification of interest TLR7, TLR8 and a housekeeping (β-actin) genes, specific forward and reverse primers provided by the Macrogen company (South Korea) were used. Details of used primers are illustrated in Table 1. mRNA expression was normalised to β-actin as an endogenous control gene. Details of reagents, volumes and conditions that are used for performing RT-qPCR amplification are showed in Tables 2 and 3. The 2−ΔΔCt method was used to estimate the relative changes in gene expression and presented as a fold-difference relative to the healthy control group. The obtained qPCR data for each gene of (TLR-7 and TLR-8) and β-actin as housekeeping genes were analysed using the ∆CT method,21 according to these steps:
ΔCT patient = CT patient − CT B-actin gene.
ΔCT control = CT control − CT B-actin gene.
ΔΔCT = ΔCT patient − ΔCT control.
Gene expression (Exp.) = 2−ΔΔCT.
Fold change (FC) = Exp. patients/Exp. Controls.
The data were categorised into two groups, treatments (vaccinated) and controls (unvaccinated). To find the effect of the COVID-19 vaccine on five cytokine concentrations and TLR7, TLR8 between the treatment and control groups, the Shapiro–Wilk test was used to assess the normality of the data. Results indicated that the treatments (vaccinated) group and controls (unvaccinated) were not normally distributed (p-value <0.05). Based on that, the Mann–Whitney U Test was used to find the statistical differences between these groups. Pearson correlation coefficients were calculated to evaluate the relationships between cytokine levels, and 95% confidence intervals (CIs) were determined to assess the precision of these correlations. All statistical analyses were done on Minitab statistical software (version 22). In the present study, (p-values <0.05) were considered significant.
ResultsSoluble cytokines levelCytokine markers are a group of polypeptide signalling molecules that can induce and regulate many cellular biological processes by stimulating cell receptors at the surface. SARS-CoV-2 can induce proinflammatory cytokines in the body. SARS-CoV-2 has been shown to be associated with activation of innate immunity and adaptive immunity. During this study, we assessed serum cytokines levels by comparing between COVID-19 vaccinated group and unvaccinated individuals. Serum concentrations of 5 cytokines showed statistically no significant differences between vaccinated participants with BBIBP-CorV vaccines and non-vaccinated individuals (p > 0.05); for IL-7, IL-10, IL-12, IL-18 and IL-21, the p-values were 0.468, 0.41, 0.257, 0.781 and 0.859, respectively (Fig. 2).
The bar graph illustrates the mean concentration (pg/ml) of cytokines in the serum of volunteers vaccinated with BBIBP-CorV COVID-19 compared to an unvaccinated control group, as measured by ELISA. Cytokine expression levels were assessed in serum using a sandwich ELISA, with two replicates per sample. The vaccinated group included (n = 60) and control group (n = 30). Non-significant differences (NS) are indicated where (p > 0.05). Data are presented as mean ± standard deviation (SD), and error bars represent the SD.
Serum sample analysis for cytokines production (IL-7, IL-10, IL-12, IL-18 and IL-21) among the vaccinated group showed that there were three types of correlation (Fig. 3A and B). Matrix plots of cytokine levels with 95% confidence intervals for Pearson correlation analysis indicated that IL-10 was positively correlated with both IL-12 and IL-21 levels (0.112, 0.397), respectively. Additionally, IL-12 correlated weakly with IL-21 (0.244). In contrast, serum cytokine levels of IL-7 levels negatively correlated with IL-12 (−0.029) and IL-21 (−0.015). Also, the level of IL-18 perfumed the same negative correlation with IL-10 (−0.094) and IL-12 (−0.036). The level of IL-7 among the vaccinated group was not correlated with IL-10 and IL-18. Also, IL-18 was not correlated with IL-21.
Correlation coefficients and patterns between the levels of different cytokines: IL-7, IL-10, IL-12, IL-18, and IL-21 in response to the BBIBP-CorV COVID-19 vaccine. A: Correlation Matrix of Cytokine Levels with 95% Confidence Intervals (CI), B: Correlogram of correlation coefficients between multiple variables. In a correlogram. The colour scale ranges from blue to red, where blue indicates a negative correlation, red indicates a positive correlation, and the intensity of the colour corresponds to the strength of the correlation. Each cell in the correlogram represents the correlation coefficient between two cytokines, with values ranging from −0.2 to 0.4. For instance, IL-10 and IL-21 show the highest positive correlation (r = 0.40), while IL-12 and IL-10 have a slight negative correlation (r = −0.09). This visualisation helps in understanding the interrelationships among these cytokines and their potential co-regulation in biological processes. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
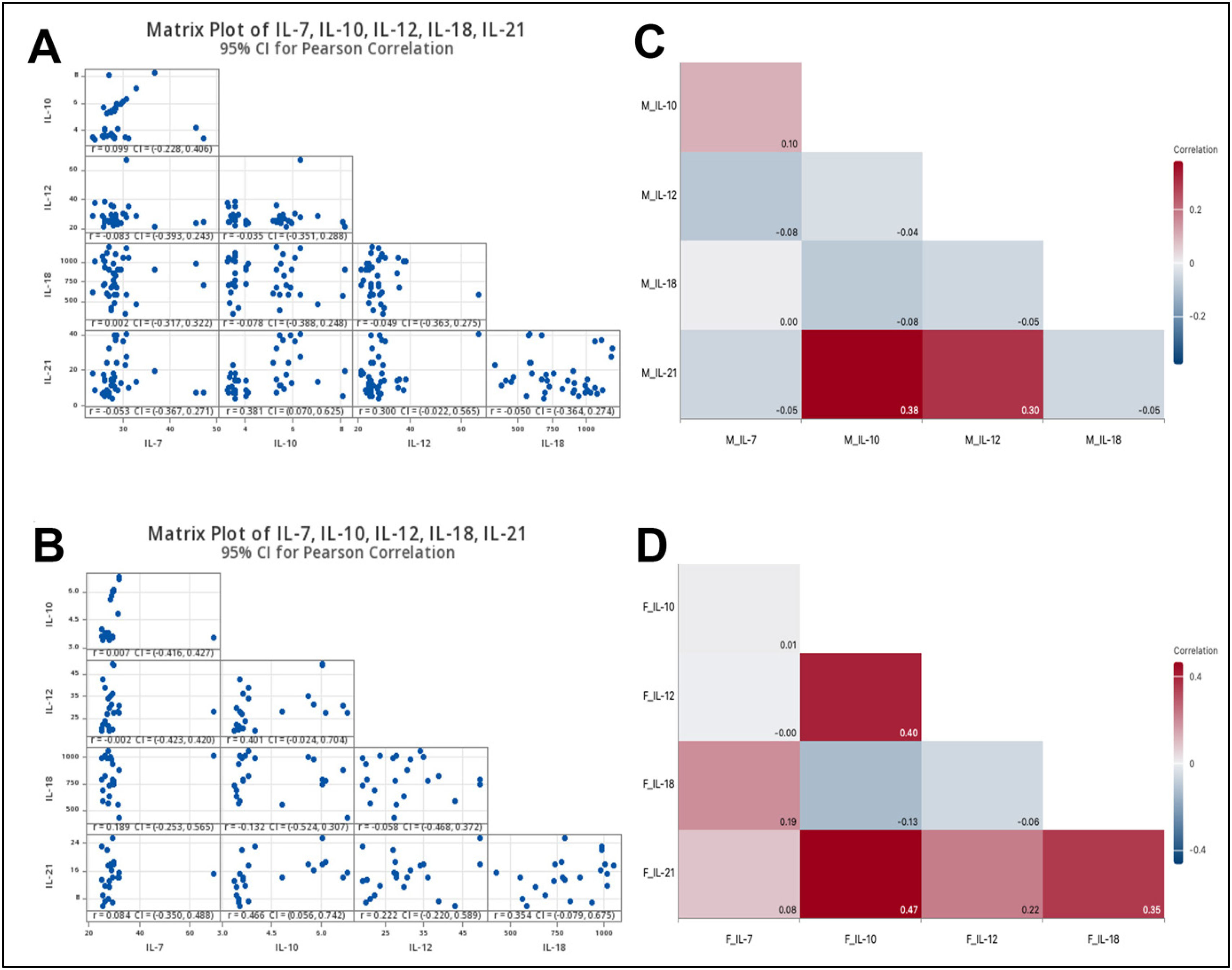
The correlations between cytokine levels based on gender within the vaccinated group were analysed. A correlation matrix and heatmap were used to visualise the relationships between cytokine levels, providing insights into how these variables interact in males (Fig. 4A and C) and females (Fig. 4B and D).
Matrix Plot and heatmap illustrate the correlation of cytokine levels in vaccinated individuals. Matrix plot of cytokine levels (IL-7, IL-10, IL-12, IL-18, IL-21) with 95% confidence intervals (CIs) for Pearson correlation in individuals vaccinated with the BBIBP-CorV COVID-19. A and C: Matrix plot and heatmap of cytokine levels in Male subjects and B and D: Matrix plot and heatmap of cytokine levels in Female subjects. The matrix displays pairwise scatter plots of cytokine levels, with Pearson correlation coefficients (r) and 95% CIs indicated. Diagonal elements represent the distribution of individual cytokine levels, while off-diagonal cells show scatter plots of pairwise cytokine relationships. The study included a total of 60 participants, comprising 38 males and 22 females. The colour scale of heatmaps represents the correlation coefficient, where red shades indicate positive correlations, blue shades indicate negative correlations, and white represents no correlation. Numerical values within each cell represent the exact correlation coefficients. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
In vaccinated males, the result showed a very weak positive correlation was observed between IL-10 and IL-7, with a Correlation coefficient (r) = 0.099, the wide 95% CI = (−0.228, 0.406), the correlation was not statistically significant (p > 0.05). A moderate positive correlation was noticed between IL-21 and IL-10,r = 0.381, 95% CI = (0.070, 0.625), while IL-21 levels exhibited a weak to moderate positive correlation with IL-12, r = 0.300 and 95% CI = (0.022, 0.565) with no statistical differences (p > 0.05). Additionally, the levels of (IL-12 and IL-7),r = −0.083, 95% CI = (−0.393, 0.243); (IL-12 and IL-10), r = −0.035, 95% CI = (−0.351, 0.288); (IL-18 and IL-10), r = −0.078 and 95% CI = (−0.388, 0.240), (IL-18 and IL-12), r = −0.049 and 95% CI = (−0.363, 0.275); (IL-21 and IL-7), r = −0.053 and 95% CI = (−0.367, 0.271), and finally (IL-21 and IL-18), r = −0.050 and 95% CI = (−0.364, 0.274), were very weakly negatively correlated (p > 0.05). Moreover, the serum level of IL-18 was not correlated with IL-7, r = 0.002 and 95% CI = (−0.317, 0.322), as there were no significant differences (p > 0.05). Together, most correlations between serum cytokine levels in males were weak and not statistically significant. The only statistically significant correlation in males was between IL-21 and IL-10 (Fig. 4A and C).
In vaccinated females, based on pairwise Pearson Correlations analysis, the only significant correlation was observed between IL-21 and IL-10 (p < 0.05), indicating a moderate positive relationship, r 0.466 and 95% CI: (0.056, 0.742), indicating that higher IL-21 levels are associated with higher IL-10 levels. The other cytokine level correlations in vaccinated females either were weak associations or were not statistically significant. Also, IL-10 levels in vaccinated females exhibited moderate correlation with IL-12, r 0.401 and 95% CI: (−0.024, 0.704). Additionally, (IL-21 and IL-18), (IL-18 and IL-7), and (IL-12 and IL-21) were weak positive correlated in females, with no statistically significant (p > 0.05). In contrast, IL-18 was negatively correlated with both IL-10 and IL-12. A very weak negative correlation was observed in females between the levels of IL-7 and IL-12. Finally, IL-7 did not correlate with IL-10 (Fig. 4B and D).
mRNA expression of TLRsEvaluation primer specific bindingThe results of evaluation results of the primer's specific binding of the TLR7 and TLR8 genes, SYBR Green chemistry was used, and the specific binding to TLRs genes fragment as the melting curve showed one peak for all samples showed in Fig. 5A–C. TLR7, TLR8 and β-actin melting and amplification curves are illustrated in Fig. 5D–F. Gel electrophoresis data of amplified DNA bands, sizes, and the number of base pairs compared with the 100 bp DNA ladder indicated precise amplification for the target genes (Fig. 5G–I). The quality of the samples ranged from 1.8 to 1.9 A260/A280, and the concentrations of RNA samples ranged from 35 to 113 ng/μl. The result of RT-PCR analysis of genes encoding TLR7 and TLR8, including β-actin as a housekeeping gene, revealed a notable upregulated expression of the target genes in the vaccinated individual compared with the control group (un-vaccinated) with statistical differences (p < 0.05) (Fig. 5J and K). The mRNA expression of TLR7 (fold changemean = 3.26, p = 0.047) and TLR8 (fold change mean = 2.35, p = 0.024) in vaccinated group compared with the un-vaccinated group TLR7 (fold changemean1.82) and TLR8 (fold changemean 1.35) was statistically significantly upregulated.
Gene expression of TLR 7 and TLR8 of vaccinated subjects with BBIBP-CorV COVID-19 compared to an unvaccinated control group. A: The melting curve of TLR7 in several samples. B: The melting curve of TLR8 in several samples. C: The melting curve of β-actin as a housekeeping gene in several samples. The specific binding of primers with target genes showed one peak. D: Amplification curves of TLR7; E: Amplification curves of TLR8; F: Amplification curves of housekeeping β-actin genes using SYBR Green chemistry. G: Gel electrophoresis of TLR7 gene expression DNA band (144 bp); H: Gel electrophoresis of TLR8 gene expression DNA band (145 bp); I: Gel electrophoresis of housekeeping β-actin gene expression DNA band (150 bp) compared with the standard DNA ladder. Gene expression levels of TLR7 (J) and TLR8 (K) were measured in peripheral blood samples from individuals vaccinated with BBIBP-CorV COVID-19 vaccine and an unvaccinated control group. The mean fold changes in gene expression are displayed to highlight differences between the groups. Statistically significant variations (p < 0.05) were determined using the Mann–Whitney U test, with p-values indicated. Data are presented as mean ± standard deviation (SD), and error bars represent the SD. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Immune response for both arms and innate and adaptive immunity plays a pivotal role in SARS-CoV-2 infections. Cytokine markers are a group of signalling molecules that can stimulate and regulate cellular activity through cell receptors at the surface. Viral infections by SARS-CoV-2 activate the cellular innate immunity phagocytosis by neutrophils and mononuclear phagocytes.25 The intersection of immunology and genetics has broadened our understanding of how the immune system responds to infection and vaccination. The immunogenetic aspects of the response to the SARS-CoV-2 infection are currently being extensively studied to reveal the understanding of this viral infection and manage the development of COVID-19 vaccines.
Cellular adaptive immune response components, CD4+ and CD8+ T cells, are important in promoting antibody production by B cells via T-cell depending manner and mediate activation of CD8+ T cells to lysis of SARS-CoV-2 infected cells.26 Despite the success of the international vaccination programme, adverse events following vaccination and the mechanisms behind them still need to be better understood. To estimate the safety of COVID-19 vaccination, Here, we evaluate the level of some cytokines following receipt of the first dose of COVID-19 vaccine as some reports indicated COVID-19 vaccine association associated with adverse events including death.27–29
In this study, we measured the cytokine responses and TLR7 and TLR8 gene expression in individual serums after the first dose of immunisation after the first dose of the BBIBP-CorV COVID-19 vaccine. Our results revealed that there were no significant differences in levels of IL-7, IL-10, IL-12, IL-18 and IL-21 cytokines between the vaccinated group compared to the unvaccinated participants. Our findings align with the previous studies examining the inflammatory markers seven to fourteen days after BNT162b2 vaccination, which found no significant evidence for increased inflammatory markers associated with vaccination at the observed time intervals.30
Some studies reported the same trend of returning some cytokines and chemokines to baseline level by less than a week after vaccination with BNT162b2,31 less than the timepoint investigated in our study. Similarly, the same transient response was found in the elevation in cytokine and chemokine levels, mice sera, and returning to baseline values by day 3.32 Another finding from this study suggested that the IL-7, IL-10, IL-12, IL-18, and IL-21 cytokines behave normally as they hit the normal value after 60 days of immunisation compared with the control group. This suggested that the innate and adaptive immune system works properly after the BBIBP-CorV COVID-19 vaccination without leading to side effects such as production abnormalities of IL-7, IL-10, IL-12, IL-18, and IL-21 when compared to the control group.
Many works studied the role of TLR gene expression, such as TLR3, TLR7 and TLR8, as endosomal innate immune sensors that are able to detect ssRNA of the SARS-CoV-2.13,33 These receptors are involved in inducing the production of Type1 IFN and the release of other proinflammatory cytokines that may be critical for the virus spread.3 The impact of COVID-19 vaccines, particularly those related to BBIBP-CorV COVID-19, was not studied. To our knowledge, this study represents the first global effort to comprehensively investigate the effects of the BBIBP-CorV COVID-19 vaccine on the human immune response.
The TLR gene expression results showed an upregulation in TLR7 and TLR8. There were significant differences between these receptors within the vaccinated group by inactivated BBIBP-CorV COVID-19 vaccine after 60 days compared with un-vaccinated group vaccination. This result suggests that this kind of vaccine evokes the immune system by presenting the SARS-Cov-2 structural proteins, allowing the immune cells to recognise and respond to this virus without causing actual infections. As a result, the immune response typically ends with the production of neutralising antibodies in addition to establishing memory T-cells and B-cells. Generally, inactivated vaccines are safer for immunocompromised individuals as the virus loses its replication ability in the host. Moreover, hitting the baseline of endosomal TLR mRNA expression refers to the safety effects of the BBIBP-CorV COVID-19 vaccine.
LimitationsThis study has several limitations. First, the sample size was relatively small and not fully representative of diverse populations, which may limit the generalizability of the findings. Second, the analysis was restricted to a limited number of cytokines, potentially overlooking other important immune markers that could provide additional insights into the vaccination response. Third, cytokine levels and TLR gene expression were measured only at a single time point, 60 days post-vaccination, without repeated measurements or evaluations during the follow-up period. This limits our ability to assess the dynamic changes in immune responses over time. Future studies should address these limitations by including larger, more diverse cohorts, expanding the range of cytokines analysed, and incorporating longitudinal designs with multiple time points to better understand the temporal dynamics of immune responses following vaccination.
ConclusionsIn summary, this study highlights the safety and efficacy of the BBIBP-CorV COVID-19 vaccine in eliciting a robust immune response without significant adverse effects. Our findings indicate that the vaccine does not lead to abnormal cytokine production, maintaining normal levels of key cytokines such as IL-7, IL-10, IL-12, IL-18, and IL-21. Additionally, the observed upregulation of TLR7 and TLR8 gene expression in vaccinated individuals suggests that the vaccine effectively stimulates the innate immune response by enabling immune cells to recognise and respond to SARS-CoV-2 antigens without causing infection. However, it is important to note that the measurements in this study were conducted only once, 60 days after vaccination, which limits our ability to assess long-term immune dynamics. Despite these limitations, this study serves as a value foundation for a pilot investigation. Future research should include longitudinal assessments with multiple time points, larger and more diverse cohorts, broader immune profiling including cytokines and cellular immune response.
Informed consent statementInformed consent was obtained from all subjects involved in the study.
FundingUniversity of Basrah, 2022.
CRediT authorship contribution statementAli Mohammed Ashraf: Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Writing – original draft. Marwan Y. Al-Maqtoofi: Conceptualization, Project administration, Visualization, Funding acquisition, Resources, Surpervision, Writing – original draft, Writing – review & editing. Ahmed A. Burghal: Conceptualization, Resources, Supervision, Validation, Writing – review & editing.
The authors declare no conflict of interest.
The authors acknowledge the participation and specimen donation in this study. We acknowledge all members of the University of Basrah, College of Science, Department of Biology, for their support.