No phenotypic methods are available to unequivocally differentiate species within the Candida glabrata complex.
AimsTo develop a new multiplex PCR method to differentiate between the three species of the C. glabrata species complex, as well as using it to study a C. glabrata collection to discover strains of the newly described species.
MethodsThe method was developed based on the Internal Transcribed Spacer (ITS) sequence differences between the species. It was validated by using a blinded collection of strains and, finally, the new molecular method was used to study a collection of 192 C. glabrata species complex strains. The obtained results were compared with ITS sequencing.
ResultsThe proposed method showed 100% concordance with ITS sequencing and proved to be effective for clinical and epidemiological applications. Two Candida bracarensis and three Candida nivariensis were found out of the 192 studied strains (0.93% and 1.40% prevalence, respectively).
ConclusionsA fast, inexpensive, robust and highly reproducible multiplex PCR method is presented. Its usefulness is demonstrated by studying a large collection of C. glabrata sensu lato strains.
No hay métodos fenotípicos disponibles para diferenciar las especies del complejo Candida glabrata.
ObjetivosDiseñar un método de PCR multiplex para diferenciar las tres especies del complejo C. glabrata y usarlo para estudiar una colección de cepas identificadas anteriormente como C. glabrata.
MétodosEl método fue desarrollado con base en las diferencias de la secuencia internal transcribed spacer (ITS) entre las especies. El método se validó mediante el uso de una colección de cepas incógnitas y se utilizó posteriormente para estudiar una colección de 192 cepas. Los resultados se compararon con las secuencias ITS.
ResultadosEl método propuesto mostró 100% de concordancia con la secuenciación de las regiones ITS y demostró ser eficaz clínica y epidemiológicamente. Se identificaron dos aislamientos de Candida bracarensis y tres de Candida nivariensis dentro de las 192 cepas identificadas fenotípicamente como C. glabrata (prevalencia de 0,93% y 1,40%, respectivamente).
ConclusionesPresentamos un método de PCR múltiplex rápido, económico y fiable. La utilidad de la metodología queda demostrada con el estudio de una gran colección de cepas de C. glabrata sensu lato.
Candida nivariensis, Candida bracarensis and Candida glabrata sensu stricto are the human pathogenic species grouped in the C. glabrata species complex of the Nakaseomyces clade.1,7,11 There are phenotypic-based methods able to differentiate these species; however, molecular methods are needed to unequivocally confirm the identification.2,3,10 The reported differences in virulence and antifungal susceptibility between the species of this complex5,13,15,21 drove the design of various molecular methodologies to identify them.6,9,12,14,18,19,22 Each of these methods has benefits and weaknesses, and none is ideal in terms of equipment costs and/or speed. The objective of this work was to propose a new multiplex PCR method designed to differentiate C. glabrata sensu stricto, C. nivariensis and C. bracarensis. The method was validated by using a blinded C. glabrata complex DNAs set. Afterwards, the multiplex PCR was used to evaluate the prevalence of C. nivariensis and C. bracarensis in a collection of 192 C. glabrata complex strains.
The strain collection, conserved in the Mycology and Molecular Diagnostics Laboratory (LMDM), included strains isolated in different Argentinian cities (Rosario, Santa Fe, Buenos Aires and Paraná) between 2001 and 2015. Each strain represents a unique isolate per patient. All the isolates were the causative agent of a proven fungal infection.8
Isolates were identified at LMDM by the assimilation and fermentation of carbohydrates, growth at different temperatures, colony color and morphology on CHROMAgar Candida®, and other morphological features.3,16,17 Identifications were confirmed by sequencing the 5.8S RNA gene and adjacent internal transcribed spacer 1 and 2 (ITS1, ITS2) regions.23 This last procedure was considered the “gold standard” when the specificity of the newly proposed multiplex PCR method was determined. The ITS region sequences were compared with those published for C. glabrata sensu stricto CBS 138, C. nivariensis CBS 9983 and C. bracarensis CBS 10154 (GenBank accession no. AY198398, GU199443.1 and GU199440.1, respectively).
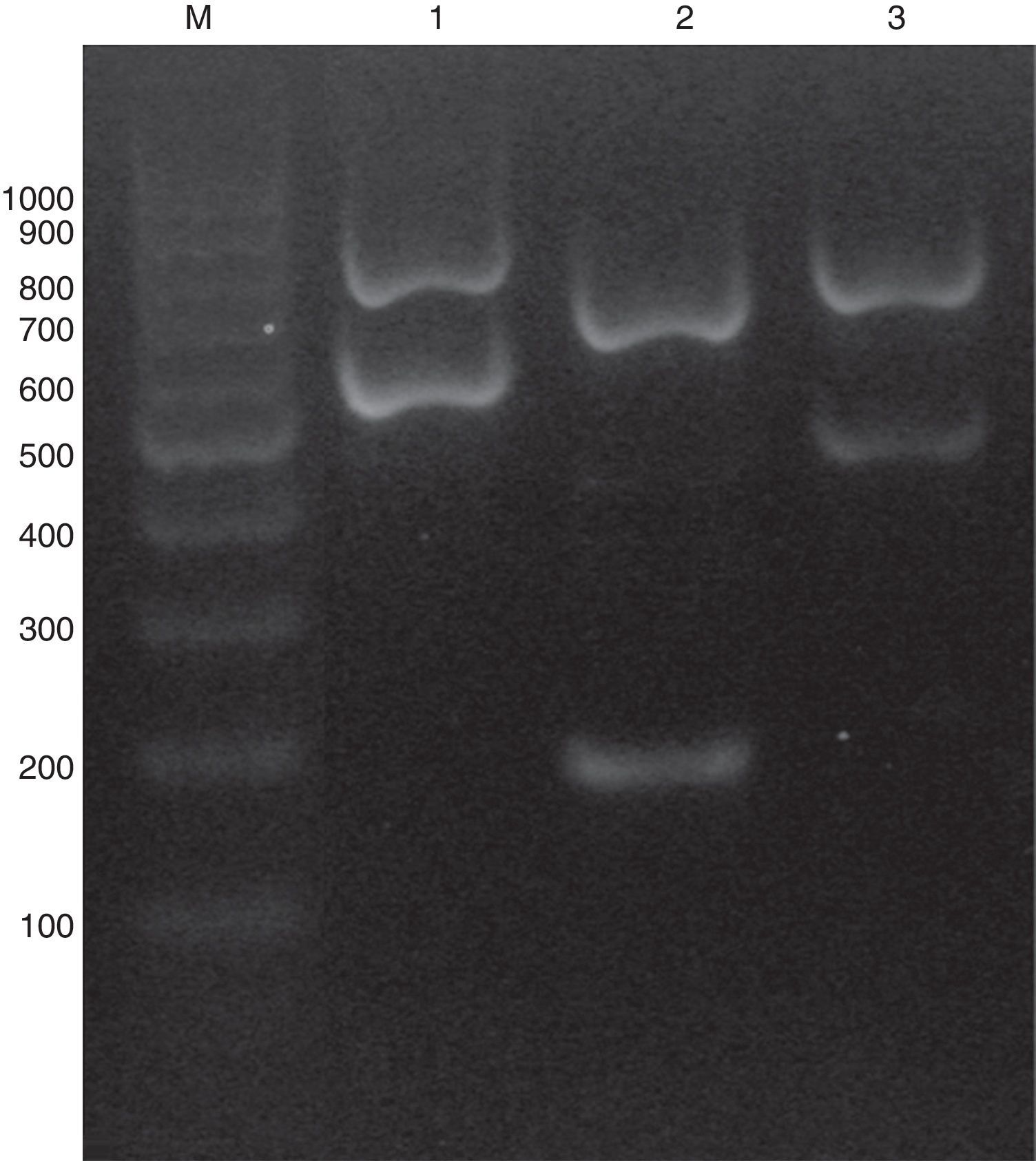
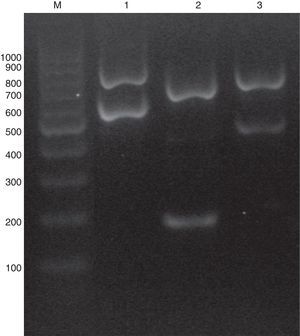
C. glabrata sensu lato genomic DNAs were extracted using phenol–chloroform method.20 A multiplex PCR was used combining five primers: ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′), ITS4 (5′-TCCTCCGCTTATTGATATGC-3′), CgITS1F (5′-TTATCACACGACTCGACACT-3′), CbITS1F (5′-TATTTACAAACTTTGTCAGAAC-3′) and CnITS2F (5′-ATGCGGACGTGCATGGTG-3′). ITS1 and ITS4 are the universal primers used for ITS regions amplification,23 while CgITS1F, CbITS1F and CnITS2F were designed to specifically hybridize C. glabrata sensu stricto ITS1, C. bracarensis ITS1 and C. nivariensis ITS2 regions, respectively. Primers were purchased from Integrated DNA technologies (IDT-Biodynamics, Buenos Aires, Argentina). PCR reactions were performed in a 25μl volume following the Pegasus DNA polymerase (PBL, Buenos Aires, Argentina) manufacturer's instructions. Briefly, each PCR tube contained 1X PCR buffer, 2mM MgCl2, 250μM dNTPs, 1 unit of Pegasus Taq polymerase, 10–50ng of yeast DNA, 0.1μM of ITS1 primer, 0.2μM of ITS4 and 0.4μM each of CgITS1F, CbITS1F and CnITS2F primers. PCRs were carried out in an Applied Biosystems thermocycler (Tecnolab-AB, Buenos Aires, Argentina) for one initial step of 2min at 94°C followed by 25 cycles of 30s at 95°C, 30s at 55°C, and 1min at 72°C, and then a final cycle of 10min at 72°C. PCR products were analyzed by electrophoresis in a 1.2% agarose gel run at 60V for one hour. When the DNA template belonged to a C. glabrata sensu stricto strain, the PCR amplification yielded two PCR amplicons (882 and 636 pb). Conversely, if the strain was C. nivariensis or C. bracarensis different band patterns were seen: 760 pb/220 pb and 805/521 pb, respectively (Fig. 1).
As stated before, the validation of the multiplex PCR was made by studying a blinded set of DNAs obtained from 30 C. glabrata complex strains (which included one C. bracarensis and two C. nivariensis) that was assembled at one of the participant Santa Fe city hospitals. There, code numbers were assigned. All the DNAs of the blinded set were correctly classified by the multiplex PCR method. Afterwards, when the strain collection was studied using the multiplex PCR, we found that most of the strains were C. glabrata sensu stricto and only two were identified as C. bracarensis and three as C. nivariensis (0.93% and 1.40% prevalence, respectively). When the multiplex PCR results were compared with ITS sequencing, there were no discrepancies (100% specificity).
Since the description of the C. glabrata complex some authors proposed differentiation methodologies based on morphology and colony color features.1,3,17 It was rapidly clear that these methods were not 100% specific since some results overlapped.3,17 Later, it was confirmed that commercial identification systems (e.g. VITEK 2) misclassified all the members of the complex as C. glabrata sensu stricto.1,3,7,17 Therefore, other methods were proposed including a peptide nucleic acid fluorescence in situ hybridization (PNA-FISH),4 a touchdown PCR differentiation based on the amplicon size of the RPL31 gene,9 a restriction enzyme fingerprinting,6 PCR coupled with dHPLC,22 MALDI-TOF12 and a differentiation method based on RFLP.18
Herein, we present a multiplex PCR method for the identification of the species of the C. glabrata complex with 100% concordance with ITS sequencing. This methodology is fast, inexpensive and it uses standard PCR equipment making it accessible for a big number of laboratories. Nevertheless, it has the same disadvantages as any classical PCR procedure: the need of electrophoresis. The main difference between this newly proposed method and the others is that we demonstrate its reliability using a blinded DNAs set while its utility was confirmed using a large collection of C. glabrata sensu lato strains.
This study was supported by Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), grant PIP2011/331 to G.G.E., and by Universidad Nacional del Litoral (UNL), grant (CAI+D) to G.G.E. and S.G. C.D. and F.L. have a fellowship from CONICET (Argentina). D.M. has a fellowship from MinCyT (Argentina). M.S.C. has a postdoctoral fellowship from CONICET.