The prevalence of coccidioidomycosis in endemic areas has been observed to increase daily. To understand the causes of the spread of the disease and design strategies for fungal detection in clinical and environmental samples, scientists have resorted to molecular tools that allow fungal detection in a natural environment, reliable identification in clinical cases and the study of biological characteristics, such as reproductive and genetic structure, demographic history and diversification. We conducted a review of the most important molecular markers in the epidemiology of Coccidioides spp. and the diagnosis of coccidioidomycosis. A literature search was performed for scientific publications concerning the application of molecular tools for the epidemiology and diagnosis of coccidioidomycosis. The use of molecular markers in the epidemiological study and diagnosis of coccidioidomycosis has allowed for the typing of Coccidioides spp. isolates, improved understanding of their mode of reproduction, genetic variation and speciation and resulted in the development specific, rapid and sensitive strategies for detecting the fungus in environmental and clinical samples. Molecular markers have revealed genetic variability in Coccidioides spp. This finding influences changes in the epidemiology of coccidioidomycosis, such as the emergence of more virulent or antifungal resistant genotypes. Furthermore, the molecular markers currently used to identify Coccidioides immitis and Coccidioides posadasii are specific and sensitive. However, they must be validated to determine their application in diagnosis.
This manuscript is part of the series of works presented at the “V International Workshop: Molecular genetic approaches to the study of human pathogenic fungi” (Oaxaca, Mexico, 2012).
Se ha descrito un aumento constante de la prevalencia de coccidioidomicosis en zonas endémicas. Para conocer las causas de esta expansión de la enfermedad y planificar estrategias para la detección del hongo en muestras clínicas y ambientales, se ha recurrido al uso de instrumentos moleculares que permitan la detección de estos hongos en su ambiente natural, su identificación fiable en los casos clínicos y el estudio de sus características biológicas, historia demográfica, diversificación y estructura reproductora y genética. El presente estudio representa una revisión de las implicaciones más importantes de los marcadores moleculares en la epidemiología de Coccidioides spp. y el diagnóstico de la coccidioidomicosis. Para ello, se efectuó una búsqueda de los artículos publicados sobre la aplicación de los instrumentos moleculares en la epidemiología y el diagnóstico de la coccidioidomicosis. El uso de marcadores moleculares en el estudio de la epidemiología y el diagnóstico de la coccidioidomicosis ha permitido tipificar aislamientos de Coccidioides spp., conocer su modo de reproducción, variabilidad genética y su especiación, así como la planificación de estrategias más rápidas, específicas y sensibles para detectar el hongo en muestras ambientales y clínicas. Los marcadores moleculares han revelado la variabilidad genética de Coccidioides, hallazgo importante porque puede influir en la epidemiología de la coccidioidomicosis, como la aparición de genotipos más virulentos o resistentes a los antimicóticos. Por otro lado, los marcadores moleculares para la identificación de Coccidioides immitis y Coccidioides posadasii, descritos hasta la fecha, son específicos y sensibles; sin embargo, deben validarse para determinar su aplicación en el diagnóstico.
Este artículo forma parte de una serie de estudios presentados en el «V International Workshop: Molecular genetic approaches to the study of human pathogenic fungi» (Oaxaca, México, 2012).
The genus Coccidioides consists of two closely related species, Coccidioides immitis and Coccidioides posadasii. Both species have been classified in the Onygenales order and Ascomycota phylum. Coccidioides is a dimorphic pathogen that grows as a filamentous saprobe in soil when not infecting mammals. This phase is very resistant, which allows the fungus to remain viable in arid or semi-arid soils for months or years. Humid soil favors fungal growth, which gives rise to the formation of asexual propagules (arthroconidia). The wind disperses the arthroconidia and they can later be inhaled by humans or animals.15,28 Once in the host, the arthroconidia become spherules, which is a morphological change of polarized isotropic growth. Spherules subsequently differentiate to produce internal spores (endospores). When the spherules rupture, the endospores are released and are able to propagate in the host and restart the spherulation cycle, a process that may result in systemic fungal infection in humans and other vertebrates.4 Previously, C. immitis was the only known causal agent of coccidioidomycosis. However, after 2002, based on microsatellite analysis, it was concluded that this species includes two different taxa: C. immitis, which is endemic in California and C. posadasii, which is endemic in the southwestern part of the United States of America and northern Mexico, as well as Central and South America.16 Currently, an increasing number of outbreaks have occurred in endemic areas, which has generated interest in studying these fungi and their genetic and reproductive characteristics, demographic history and diversification process. Depending on the purpose of study, several molecular markers, alone or in combination, have been used in the development of specific and sensitive strategies for the diagnosis of coccidioidomycosis.
Molecular markersMolecular markers are a dataset collected from various molecular techniques and represent unique genetic traits in individuals, populations or species. Variants of these genetic traits are the result of randomly occurring mutations or mutations influenced by the environment. The latter results in a genetic variation (polymorphism) and is essential for the adaptation to environmental changes and, therefore, the survival of the species.24
In order to detect genetic differences in the molecular markers, several properties must be present: whether variable or polymorphic, the genetic change must have a defined inheritance pattern, be observed frequently in the genome, occur under neutral selection (not influenced by the environment), be easily accessed, and have high reproducibility. Because it is extremely difficult to find a molecular marker that meets all the above criteria, some authors use combinations of different molecular markers, as they are most informative.24
Molecular markers have been used in the study of coccidioidomycosis to identify species, type isolates, determine the structure or reproductive mode and identify the degree of genetic differentiation between isolates and species. At the clinical level, knowledge of the genetic variability of the Coccidioides species, allows for the development of antifungal agents and vaccines, because their design will take into account genetic diversity, thus ensuring that the action of these biological products reliable covers all genotypes of the species. Furthermore, the use of molecular markers is an alternative to improve the diagnosis of coccidioidomycosis, because conventional methods for detecting and identifying the fungus are time-consuming and less sensitive and specific.
Role of molecular markers in the epidemiology of coccidioidomycosisGenetic variability of Coccidioides spp.The first studies identifying genotypic variability in populations of the Coccidioides genus only included C. immitis; C. posadasii was not formally recognized until 2002. The first genotypic method used to analyze the genetic variability and related isolates of C. immitis was described by Zimmermann et al.38 In this study, they used Restriction Fragment Length Polymorphic (RFLP) from DNA obtained from 15 patient-isolates in California. Their results showed a similar RFLP pattern in 13 of the 15 isolates and a second pattern in the remaining two isolates, which indicated genetic diversity between the two isolates and the remainder of the isolates studied. RFLP involves obtaining DNA fragments by restriction endonuclease digestion. These fragments may vary in size and number, enabling the observation of polymorphisms among the isolates.
Furthermore, Burt et al.9 studied the pattern of genetic variation in 25 isolates of C. immitis from a hospital in Tucson, Arizona, using PCR amplification and Single-Strand Conformational Polymorphism (SSCP) detection. Their results also showed genetic variation among the examined isolates. In SSCP, double-stranded fragments are denatured by heating and then cooled to prevent further association. Single chain molecules of DNA can form secondary structures due to internal base pairing. These differences in secondary structure cause the DNA strands to migrate differently during electrophoresis with non-denaturing acrylamide. The variation in electrophoretic mobility of single-stranded DNA is likely due to changes caused by nucleotide substitutions. The variable bands are extracted from the gel and sequenced. When polymorphic regions are found, new oligonucleotides are designed and used to amplify the fragments from each isolate studied.13
In addition, Burt et al.11 utilized DNA Multilocus Sequence Typing (MLST) of C. immitis isolates from Arizona, California and Texas, to obtain information about the structure of fungal populations. There was evidence of genetic differentiation among the three populations studied, which suggests a very low level of gene flow between them. MLST involves the PCR amplification of fragments (450–500bp) of various housekeeping genes (7–8 genes), followed by sequencing both strands (alleles) to observe sequence changes. Analysis of changes in the fungal housekeeping genes allows for typing populations or isolates based on their allelic profiles.1
Recently, Sharpton et al.32 and Neafsey et al.25 used Whole Genome Shotgun (WGS) sequencing of C. immitis and C. posadasii isolates and showed that they possess significant genetic variation in their populations. The WGS approach, in which the whole genome is sheared into millions of fragments that are sequenced and reassembled to produce a series of sequence “scaffolds”, has been used to sequence the genome of various organisms.
C. immitis reproductive modeEarly investigations into the structure of recombinant C. immitis was provided by Burt et al.,9 who used Sequencing With Arbitrary Primer Pairs (SWAPP) in conjunction with SSCP, to study C. immitis isolates from Tucson, Arizona. Their results revealed genetic variability in the studied C. immitis isolates, which supports the existence of a sexual recombination process in this fungus. The SWAPP technique is a variation of Random Amplified Polymorphic DNA (RAPD). SWAPP uses two different 20bp oligonucleotides, which are aligned at low stringency temperatures. Bands that do not show any variation in agarose gel electrophoresis are extracted to perform SSCP, as described above.10
Furthermore, Koufopanou et al.,22,23 by sequencing five nuclear gene fragments from a sample of 17 C. immitis strains and analyzing the genealogy of each of the five loci, showed a minimum length between them, which is indicative of no recombination. However, when the genealogy of the loci was analyzed together, evidence of recombination was found. To acquire partial gene sequences, PCR is required and oligonucleotides are used to amplify the partial gene regions of a set of isolates under study. Oligonucleotides are designed from the GenBank sequences for the fungus of interest or a close relative or by using SSCP or Sequenced Characterized Amplified Region (SCAR) markers.18 The advantage of this method is that it achieves the highest possible resolution when obtaining the genetic information.
Other work, developed by Fisher et al.,17 focuses on elucidating the genetic diversity and the mode of reproduction of C. immitis isolates. They used two types of genetic markers, Single Nucleotide Polymorphisms (SNPs) and Short Tandem Repeats (STRs). Their results showed wide multilocus genotypic diversity for two of the loci studied. The population structure was consistent with a recombinant population. A SNP is a DNA sequence variation that affects a single nucleotide in the genome. SNPs exist throughout the genome and are abundant, particularly in the human genome, with a rate of one SNP per 1000 base pairs.37 Most SNPs are located in noncoding regions and do not have a direct impact on the phenotype of an individual. However, some SNPs introduce mutations in expressed sequences or in the regions that influence gene expression (promoters, enhancers) and can induce changes in the structure or regulation of proteins. Thus, SNPs have the potential to detect functional genetic variation. Alternatively, microsatellites, SSRs or STRs consist of a length of DNA of a few nucleotides that are repeated several times in tandem and are scattered throughout the genomes of eukaryotes. They are often displayed as hypervariable regions and usually show tens of alleles in a locus that differs in the number of repeats. Microsatellites are relatively small and therefore can be easily PCR-amplified using DNA extracted from various sources. Polymorphisms can be detected on a sequencing gel and the availability of automated DNA sequencers allows for ultrafast analysis of a large number of samples.14
In addition, Fisher et al.16 used SNPs, the partial sequences of genes and microsatellites to analyze a considerable number of Coccidioides spp. isolates from various geographical regions and showed the separation of the two taxa of Coccidioides corresponding to two phylogenetic species, which supports previous findings. They also confirmed that C. posadasii represents a monophyletic recombinant clade.
Speciation of Coccidioides spp.The studies carried out by Koufopanou et al.,21–23 who used partial sequences of known genes, were the first to demonstrate the existence of cryptic species in the Coccidioides genus. Moreover, Fisher et al.,16 using microsatellites as mentioned above, confirmed the separation of the two taxa of the Coccidioides genus, which correspond to two phylogenetic species. Moreover, the results of the phylogenetic analysis defined the separation of the two species. For isolates from Californian endemic areas, the name of C. immitis was reserved, and isolates from the endemic areas of Arizona, Texas, Mexico and South America were referred to as C. posadasii.
Undoubtedly, the application of various molecular markers to studies on the genetic variability of C. immitis and C. posadasii, demonstrated that both species show wide genetic variability, most likely due to a process of sexual recombination, for which there is increasingly compelling evidence in these fungi. Moreover, this variability is becoming more important as new genotypes may appear, possibly with increased virulence or antifungal resistance, which may directly affect the management of patients with coccidioidomycosis and explains the increased number of disease cases in endemic areas and non-endemic areas. Thus, it is important to conduct further studies on the genetic variation of these species that contributes to the epidemiology of the disease.
Role of molecular markers in the diagnosis of coccidioidomycosisLaboratory methods for the diagnosis of fungal infections use three broad approaches: microscopic detection of the etiologic agent in clinical material, the isolation and identification of the fungus in cultures, and the detection of a serological response to the pathogen or a marker of their presence, such as a fungal cell constituent or metabolic product. In recent years, new diagnostic procedures based on the detection of fungal DNA in clinical material have been developed; however, these techniques have not yet had a significant impact in most clinical laboratories.
Molecular techniques for the detection and identification of Coccidioides spp. can be divided into two types: (a) signal amplification methods using nucleic acid hybridization, which the hybridization probes can be used either to confirm the identification of a culture or to identify the fungi in tissue sections, and (b) nucleic acid amplification, which includes all PCR-based techniques.2 The hybridization methods use chemiluminescent probes, such as the Accuprobe and Diversi-Lab systems commercially available from Gen-Probe, Inc. (San Diego, CA) and Bacterial Barcodes, Inc. (Athens, GA), respectively and have been proposed for detecting Coccidioides and other fungi. The PCR-based techniques are a result of specific research with the fungus and these offer the greatest potential sensitivity and specificity for the detection and identification of Coccidioides.
For PCR, a wide range of targets have been used, highlighting specific Coccidioides genes, such as the genes Ag2/PRA, CSA and rRNA genes (18S, 28S, 5.8S rRNA genes), but the latter are the most frequently used because of their universal nature and large copy number, which gives them greater sensitivity. The PCR conditions used to detect Coccidioides species are single, multiplex, nested, semi-nested, real-time and PCR-RFLP (Table 1). These procedures are distinctive because of their specificity and sensitivity, which are adequate for properly diagnosing coccidioidomycosis and detecting the pathogen in environmental samples. However, implementation of these techniques in hospital laboratories has not yet begun. These techniques have not been validated in coccidioidomycosis-endemic areas because the reproducibility between laboratories is unknown. Another aspect which has hampered their implementation is that only a few of these methods are commercially available, and some involve laborious methods, such as PCR-RFLP. In most clinical laboratories, experienced personnel are required, which makes these molecular tools accessible only in research laboratories. Therefore, the use of these tools must become widespread so that each clinical laboratory can decide how to integrate molecular methods into their standard identification practices.29,30
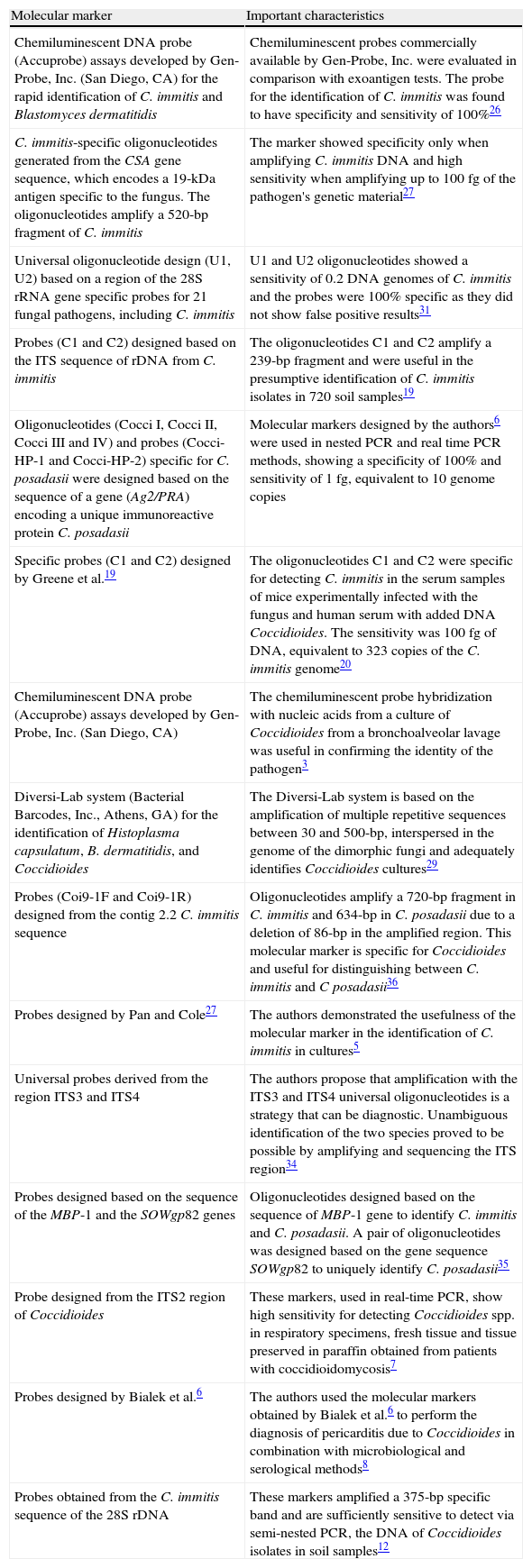
Molecular markers used in the diagnosis of coccidioidomycosis.
| Molecular marker | Important characteristics |
| Chemiluminescent DNA probe (Accuprobe) assays developed by Gen-Probe, Inc. (San Diego, CA) for the rapid identification of C. immitis and Blastomyces dermatitidis | Chemiluminescent probes commercially available by Gen-Probe, Inc. were evaluated in comparison with exoantigen tests. The probe for the identification of C. immitis was found to have specificity and sensitivity of 100%26 |
| C. immitis-specific oligonucleotides generated from the CSA gene sequence, which encodes a 19-kDa antigen specific to the fungus. The oligonucleotides amplify a 520-bp fragment of C. immitis | The marker showed specificity only when amplifying C. immitis DNA and high sensitivity when amplifying up to 100fg of the pathogen's genetic material27 |
| Universal oligonucleotide design (U1, U2) based on a region of the 28S rRNA gene specific probes for 21 fungal pathogens, including C. immitis | U1 and U2 oligonucleotides showed a sensitivity of 0.2 DNA genomes of C. immitis and the probes were 100% specific as they did not show false positive results31 |
| Probes (C1 and C2) designed based on the ITS sequence of rDNA from C. immitis | The oligonucleotides C1 and C2 amplify a 239-bp fragment and were useful in the presumptive identification of C. immitis isolates in 720 soil samples19 |
| Oligonucleotides (Cocci I, Cocci II, Cocci III and IV) and probes (Cocci-HP-1 and Cocci-HP-2) specific for C. posadasii were designed based on the sequence of a gene (Ag2/PRA) encoding a unique immunoreactive protein C. posadasii | Molecular markers designed by the authors6 were used in nested PCR and real time PCR methods, showing a specificity of 100% and sensitivity of 1fg, equivalent to 10 genome copies |
| Specific probes (C1 and C2) designed by Greene et al.19 | The oligonucleotides C1 and C2 were specific for detecting C. immitis in the serum samples of mice experimentally infected with the fungus and human serum with added DNA Coccidioides. The sensitivity was 100 fg of DNA, equivalent to 323 copies of the C. immitis genome20 |
| Chemiluminescent DNA probe (Accuprobe) assays developed by Gen-Probe, Inc. (San Diego, CA) | The chemiluminescent probe hybridization with nucleic acids from a culture of Coccidioides from a bronchoalveolar lavage was useful in confirming the identity of the pathogen3 |
| Diversi-Lab system (Bacterial Barcodes, Inc., Athens, GA) for the identification of Histoplasma capsulatum, B. dermatitidis, and Coccidioides | The Diversi-Lab system is based on the amplification of multiple repetitive sequences between 30 and 500-bp, interspersed in the genome of the dimorphic fungi and adequately identifies Coccidioides cultures29 |
| Probes (Coi9-1F and Coi9-1R) designed from the contig 2.2 C. immitis sequence | Oligonucleotides amplify a 720-bp fragment in C. immitis and 634-bp in C. posadasii due to a deletion of 86-bp in the amplified region. This molecular marker is specific for Coccidioides and useful for distinguishing between C. immitis and C posadasii36 |
| Probes designed by Pan and Cole27 | The authors demonstrated the usefulness of the molecular marker in the identification of C. immitis in cultures5 |
| Universal probes derived from the region ITS3 and ITS4 | The authors propose that amplification with the ITS3 and ITS4 universal oligonucleotides is a strategy that can be diagnostic. Unambiguous identification of the two species proved to be possible by amplifying and sequencing the ITS region34 |
| Probes designed based on the sequence of the MBP-1 and the SOWgp82 genes | Oligonucleotides designed based on the sequence of MBP-1 gene to identify C. immitis and C. posadasii. A pair of oligonucleotides was designed based on the gene sequence SOWgp82 to uniquely identify C. posadasii35 |
| Probe designed from the ITS2 region of Coccidioides | These markers, used in real-time PCR, show high sensitivity for detecting Coccidioides spp. in respiratory specimens, fresh tissue and tissue preserved in paraffin obtained from patients with coccidioidomycosis7 |
| Probes designed by Bialek et al.6 | The authors used the molecular markers obtained by Bialek et al.6 to perform the diagnosis of pericarditis due to Coccidioides in combination with microbiological and serological methods8 |
| Probes obtained from the C. immitis sequence of the 28S rDNA | These markers amplified a 375-bp specific band and are sufficiently sensitive to detect via semi-nested PCR, the DNA of Coccidioides isolates in soil samples12 |
Moreover, the use of molecular methods to identify organisms is based on the assumption that strains belonging to the same species show less genetic variation than organisms that are less closely related.33 However, this is not true for Coccidioides or other pathogenic fungi because genetic variability has been shown to exist among isolates from different geographic regions, although their sexual state has not been described. Molecular markers to be used for the detection of pathogens from clinical samples should be designed from autochthonous isolates in the region in which they are to be used.
The molecular markers for the identification of C. immitis and C. posadasii that have been described to date are specific and sensitive; however, they must be validated to determine their applicability for diagnosis. The validation of these markers is important because they have been designed using clinical isolates from a single geographic region, which is inconvenient because the genetic variability between isolates of the fungus from different countries could lead to false results, as has occurred in other fungi. The use of molecular markers to identify Coccidioides spp. will not replace the use of conventional tests but will provide fast and reliable results.
Conflict of interestsThe authors declare that they have no competing interests.
This paper is a partial fulfillment of the requirements of the Graduate Program in Biological Sciences of the National Autonomous University of Mexico (UNAM). EDE acknowledges the support from the Program in Biological Sciences. This project was funded by PAPIIT-DGAPA (IN215509-3).