Botrytis cinerea is an ascomycete with a high genetic diversity and complex population structure, as reported from several hosts and sites. However, nothing is known about its genetic diversity in Argentina.
AimsThe aim of this work is to estimate the genetic diversity of a local population of B. cinerea isolates obtained from grapevine in Argentina.
MethodsIn this work, 35 strains that had been isolated from grapevines were genotyped for the presence of transposable elements and PCR-based RFLP molecular markers. The obtained results were compared with those from a large French population of the fungus, and used to perform a population genetics analysis using the Genepop software.
ResultsAll the analysed isolates were classified as Group II (according to the most recent proposed classification) and showed a high degree of genetic diversity, with 14 different haplotypes. A significant difference in allele frequency was recorded between the local and French populations.
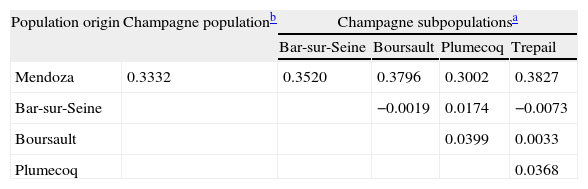
ConclusionsThese comparisons between fungal populations, led to the detection of a high level of diversity and the differentiation between local and French groups of isolates. This was confirmed by an Fst value of 0.3332, which was higher than that reported for other pairwise comparisons of populations. This work constitutes the first report on the genetic diversity of B. cinerea isolates and their population structure in Argentina.
Botrytis cinerea es un ascomiceto con una gran diversidad genética y una compleja estructura poblacional cuya presencia ha sido descrita en diversos lugares y huéspedes distintos, pero nada se sabe acerca de su diversidad genética en Argentina.
ObjetivosEl objetivo de este trabajo fue estimar la diversidad genética de una población local de aislamientos de B. cinerea obtenidos de vid en Argentina.
MétodosEn este trabajo, 35 cepas aisladas de vides fueron genotipadas mediante marcadores moleculares basados en PCR-RFLP y según la presencia de elementos transposables. Estos resultados fueron comparados con los de una gran población francesa del hongo, y utilizados para realizar un análisis de genética poblacional utilizando el software Genepop.
ResultadosTodos los aislamientos analizados fueron clasificados como grupo II (de acuerdo a la clasificación más recientemente propuesta) y mostraron un alto grado de diversidad genética, con 14 haplotipos distintos en el número de muestras involucradas. Se observó una notable diferencia en la frecuencia alélica entre ambas poblaciones.
ConclusionesEstas diferencias entre las poblaciones comparadas condujeron a la detección de un alto nivel de diversidad y diferenciación poblacional entre los aislamientos locales y los franceses. Esto fue confirmado por un valor Fst de 0,3332, superior al previamente reportado para otras comparaciones de este tipo. Este trabajo es el primero en documentar la diversidad genética de aislamientos de B. cinerea y su estructura poblacional en Argentina.
In Argentina, the area under grapevine (Vitis vinifera L.) cultivation is approximately 220,000ha, 75% of which are located in the western Mendoza province, where climate conditions are very propitious for this crop. Incidence of fungal diseases in the area is lower compared with other viticulture regions of the world. However, grey mould and bunch rot are two of the diseases that have been reported to affect grapevines in Mendoza. The fungus Botrytis cinerea Pers. (teleomorph Botryotinia fuckeliana (de Bary) Whetzel) has been shown to be responsible for the first disease and is also one of the most important causal agents of the second one (together with Aspergillus spp. and Penicillium spp.).19 Grey mould is a serious disease that limits grapevine cultivation in some regions around the world. It can cause severe losses in the quality and volume of grapes harvested for wine production or fruit market.
B. cinerea is an haploid and heteroallelic ascomycete. It has a wide host range and affects a number of plant organs. It is involved in several types of rots and is responsible for severe economic losses in vegetable, fruit and ornamental crops. B. cinerea is a complex organism, with a large genetic variability. This variation has been studied using restriction fragment length polymorphism (RFLP),7 the presence or absence of transposable elements,3,10 random amplification of polymorphic DNA (RAPD) markers,18 amplified fragment length polymorphisms (AFLP),15 and microsatellites,5,8,11 amongst other molecular typing techniques. Early population structure studies for this fungus have suggested the presence of a species complex that is based on the presence or absence of two transposable elements (boty and flipper). At that moment, two sibling sympatric species were defined: transposa, in which both elements are present, and vacuma, in which neither element is present. Isolates that contain only one element (boty or flipper) have also been reported.6,7,18 This early definition of species complex was reviewed in recent studies based on multiple gene genealogies, and a new classification with two groups of cryptic species was proposed. Group I shows a restricted host and geographic range and comprises only vacuma isolates. Group II has a wider host range, and includes the isolates that are most deleterious for grapevines and other hosts. Group II contains both vacuma and transposa isolates.4
There are no available data about the genetic variability of local isolates of the fungus, nor about the population structure of the fungus in Mendoza, but a differential response of grapevine isolates to fungicides and biocontrol agents has been reported.16,17,21
The objectives of this work were to provide information about the occurrence of genetic variants of B. cinerea, to study the genetic variability of Mendoza isolates, and to compare these findings with those from French vineyards. Considering the amount of data previously published at the time of the study, PCR-RFLP and presence of transposable elements were chosen as genotyping methods.
Material and methodsFungal isolatesA total of 35 isolates of B. cinerea were collected from vineyards of the Mendoza province, Argentina, for genetic analysis. The samples were taken from four viticulture regions (Uco Valley, South Region, High Lands of Mendoza River and East Region).1 The grapevine cultivars and geographical origins are listed in Table 1. The isolates comprised single conidial strains derived directly from the field, and were maintained in potato dextrose agar at 4°C. One foreign strain previously genotyped as vacuma was also analysed during all the evaluation process as control. This isolate was kindly supplied by Dr. Silva, from Fundación Ciencia para la Vida, Santiago de Chile.
Origin and cultivar of the 35 vineyard isolates of Botrytis cinerea included in the study.
| Isolate | Origin | Cultivar |
| 1 | Uco Valley | Pedro Giménez |
| 2 | Uco Valley | Syrah |
| 3 | East Region | Unidentified table grape cultivar |
| 4 | Uco Valley | Pedro Giménez |
| 5 | Uco Valley | Pedro Giménez |
| 6 | Uco Valley | Pedro Giménez |
| 7 | East Region | Syrah |
| 8 | East Region | Syrah |
| 9 | High Lands of Mendoza River | Pedro Giménez |
| 10 | High Lands of Mendoza River | Unidentified |
| 11 | High Lands of Mendoza River | Unidentified |
| 12 | Uco Valley | Syrah |
| 13 | High Lands of Mendoza River | Pedro Giménez |
| 14 | East Region | Unidentified |
| 15 | Uco Valley | Pedro Giménez |
| 16 | Uco Valley | Syrah |
| 17 | Uco Valley | Unidentified |
| 18 | East Region | Unidentified |
| 19 | South Region | Mixed cultivars |
| 20 | South Region | Mixed cultivars |
| 21 | Uco Valley | Pedro Giménez |
| 22 | High Lands of Mendoza River | Chenin |
| 23 | East Region | Syrah |
| 24 | East Region | Mixed cultivars |
| 25 | Uco Valley | Syrah |
| 26 | East Region | Syrah |
| 27 | East Region | Syrah |
| 28 | Uco Valley | Pedro Giménez |
| 29 | Uco Valley | Pedro Giménez |
| 30 | East Region | Unidentified table grape cultivar |
| 31 | East Region | Syrah |
| 32 | Uco Valley | Pedro Giménez |
| 33 | Uco Valley | Pedro Giménez |
| 34 | East Region | Syrah |
| 35 | South Region | Unidentified |
Genomic DNA was extracted according to the method described by Möller et al.14 Mycelia were collected from 10-day-old B. cinerea cultures grown at 25°C in the dark. The yield and integrity of the DNA were checked by agarose gel electrophoresis. The genomic DNA was used as a template for the subsequent PCR.
PCR amplification was performed using primers for the ribosomal intergenic spacer (IGS), the nitrate reductase and ATP synthase genes previously reported by Giraud et al.7 The flipper transposable element was detected by PCR using previously described primers.6 In order to detect the boty element, a new primer pair (Boty-F 5′-TAACCTTGTCTTTGCTCATC-3′ Boty-R 5′-CCCAATTTATTCAATGTCAG-3′) was designed using the sequence that corresponded to Accession Number X81791 in NCBI Genbank, using the Primer3 Software available at http://frodo.wi.mit.edu/primer3/input.htm.20 In order to validate the PCR products, the previously genotyped vacuma isolate was included in the reaction. The resulting products were checked by agarose gel electrophoresis, and the reactions repeated three times for each PCR marker prior to the RFLP analysis.
To type the isolates genetically using RFLPs, the following previously described molecular markers7 were used: the IGS amplification fragment (digested with HaeIII, BamHI, HindIII or RsaI), the ATP synthase fragment (digested with BamHI), and the nitrate reductase fragment (digested with RsaI). In addition, the IGS amplification fragment was digested with Mval. Ten microliters of the amplification products were digested directly, without further purification, by adding 3U of restriction enzyme and incubating the samples at 37°C for 2h. The restriction fragments were resolved by 2% agarose gel electrophoresis.
Data analysisThe restriction pattern for each enzyme was analysed according to Giraud et al.7 A total of eight markers were used (IGS/HaeIII, IGS/BamHI, IGS/HindIII, IGS/RsaI, ATP-S/BamHI and NR/RsaI, together with the occurrence of boty or flipper). The locus IGS/MvaI, was not considered because it was monomorphic for the local population. The samples of B. cinerea from the four different regions of the Mendoza province were considered as a single population, and were compared with those 312 samples that were collected from four different locations (Bar-sur-Seine, Boursault, Plumecoq and Trepail) in the French Champagne region and characterized previously by Giraud et al.7 using the same molecular markers. Comparisons were done with the entire French population, and with subpopulations that were defined on the basis of sampling sites in the Champagne region. In order to characterize the local population of B. cinerea, the null hypothesis H0 (the allele distribution is independent across the populations) was checked by applying population differentiation analysis and F-statistics, using the Genepop software package available at http://genepop.curtin.edu.au/. The Genepop software estimates the P value for Fisher's exact test using a Markov chain.
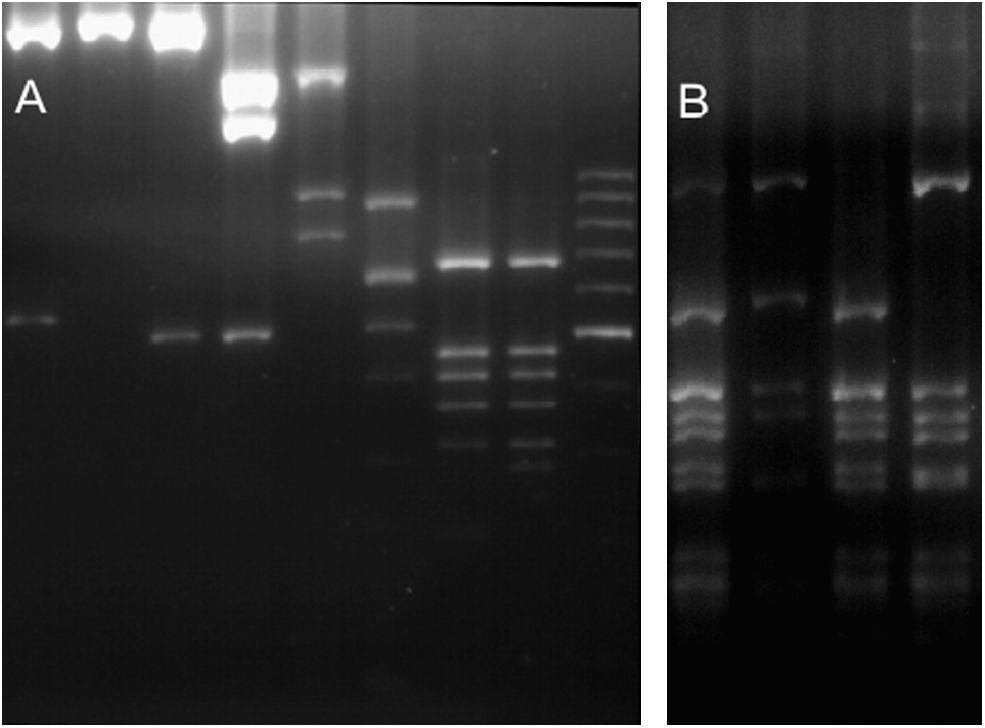
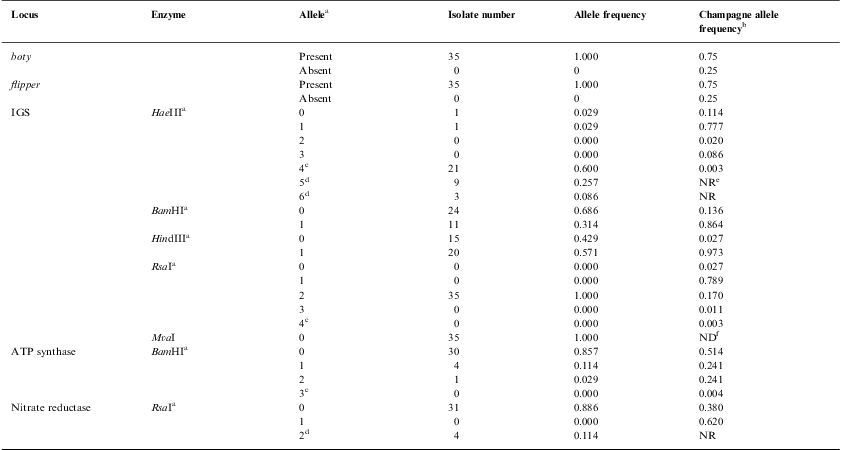
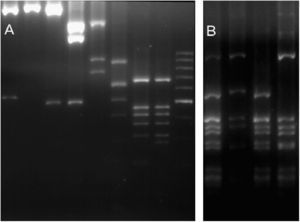
ResultsGenetic diversityFor the 36 samples that were analysed (35 local strains and the previously genotyped vacuma foreign strain), the amplified PCR products showed the expected size for all the targets tested. All the local strains contained both the boty and flipper transposable elements, and were therefore typed as transposa-type according to Giraud et al.,7 or Group II according to Fournier et al.4 Neither of the two transposable elements were amplified from the reference vacuma strain, but all the other markers were amplified from this strain. The frequency of occurrence of the different alleles is shown in Table 2, and the restriction fragment patterns for some of the different alleles are shown in Fig. 1. It should be noted that for IGS/HaeIII, most of the local isolates corresponded to alleles 4 and 5. Allele 4 has been reported as a private allele for a population,7 and allele 5 was recorded for the first time in this study of local isolates.
Percentage of each allele observed in the 35 B. cinerea local isolates analysed.
| Locus | Enzyme | Allelea | Isolate number | Allele frequency | Champagne allele frequencyb |
| boty | Present | 35 | 1.000 | 0.75 | |
| Absent | 0 | 0 | 0.25 | ||
| flipper | Present | 35 | 1.000 | 0.75 | |
| Absent | 0 | 0 | 0.25 | ||
| IGS | HaeIIIa | 0 | 1 | 0.029 | 0.114 |
| 1 | 1 | 0.029 | 0.777 | ||
| 2 | 0 | 0.000 | 0.020 | ||
| 3 | 0 | 0.000 | 0.086 | ||
| 4c | 21 | 0.600 | 0.003 | ||
| 5d | 9 | 0.257 | NRe | ||
| 6d | 3 | 0.086 | NR | ||
| BamHIa | 0 | 24 | 0.686 | 0.136 | |
| 1 | 11 | 0.314 | 0.864 | ||
| HindIIIa | 0 | 15 | 0.429 | 0.027 | |
| 1 | 20 | 0.571 | 0.973 | ||
| RsaIa | 0 | 0 | 0.000 | 0.027 | |
| 1 | 0 | 0.000 | 0.789 | ||
| 2 | 35 | 1.000 | 0.170 | ||
| 3 | 0 | 0.000 | 0.011 | ||
| 4c | 0 | 0.000 | 0.003 | ||
| MvaI | 0 | 35 | 1.000 | NDf | |
| ATP synthase | BamHIa | 0 | 30 | 0.857 | 0.514 |
| 1 | 4 | 0.114 | 0.241 | ||
| 2 | 1 | 0.029 | 0.241 | ||
| 3c | 0 | 0.000 | 0.004 | ||
| Nitrate reductase | RsaIa | 0 | 31 | 0.886 | 0.380 |
| 1 | 0 | 0.000 | 0.620 | ||
| 2d | 4 | 0.114 | NR | ||
RFLP molecular markers showing its alleles. (A) Lanes 1–2 BamHI restricted IGS (alleles 1 and 0). Lanes 3–4 HindIII restricted IGS (alleles 0 and 1). Lane 5 MvaI restricted IGS (allele 0). Lane 6 RsaI restricted IGS (allele 2). Lanes 7–8 RsaI restricted Nitrate reductase (alleles 0 and 1). Lane 9 100bp molecular weigth marker. (B) Lanes 1–4 HaeIII restricted IGS (alleles 0, 6, 5 and 4).
Fourteen different haplotypes were found among the 35 strains analysed considering the polymorphic loci. Two of these haplotypes were most frequent, being each one present in eight and seven samples (data not shown).
Population structure analysisThe genic differentiation between the Mendoza and Champagne populations showed P values of less than 0.00001 for all the loci analysed, with the exception of IGS/RsaI, which was monomorphic for the local population (P=0.00054). The IGS/Mval locus was monomorphic for the Mendoza population, and it was not compared with the French group of populations, because the information for this locus was not available. When the genic differentiation between the local population and the four individual French subpopulations was analysed, the P values were less than 0.00001 for all the other molecular markers. The population pairwise Fst values were calculated for the data sets that corresponded to the Mendoza and French populations mentioned above, and the results are displayed in Table 3.
DiscussionB. cinerea is a phytopathogenic fungus that causes severe economic losses in vegetable and fruit crops, especially grapes, in which it is responsible for grey mould in addition to other bunch rots. Several studies have reported B. cinerea vacuma and transposa as sympatric entities in several locations around the world,6,13,18 but the high prevalence of transposa strains amongst those that have been isolated from grapevines should be noted.18 In this study, we found that all of the field isolates collected from grapevines in the Mendoza region of Argentina were transposa-type (i.e. they contained both flipper and boty transposable elements). Due to the fact that no boty-only, flipper-only, or vacuma-type isolates were identified in this study, we cannot conclude that these sibling species are sympatric in Mendoza vineyards. In spite of the limited number of samples analysed, our results agree with the previous reports about the absence or very low frequency of Group I isolates amongst those that infect grapevines.8,9,11,12 A possible explanation for this observation has been suggested by Martinez et al.12 They observed that transposa isolates showed a higher virulence than vacuma isolates in grapevines, which led to a higher frequency of occurrence of transposa compared to vacuma.
Fourteen different haplotypes were identified amongst the 35 samples analysed, and it is noteworthy that two of these haplotypes accounted for 40% of the total samples. The isolates that shared the same haplotype were not related geographically. Also, in previous works, the resistance levels for two widely used fungicides was evaluated, but no correlation was found between this behaviour and the haplotypes information.16,17 Today it is well known that the resistance for Carbendazim and Iprodione fungicides is related to single nucleotide polymorphism in the b-tubulin and osmosensing histidine kinase Bos-1 genes, respectively.2,22
As stated by Giraud et al.7 if two populations do not exchange genetic material for a long period of time, the allele frequency may vary between them. A proof of this variation is shown in Table 2, where a significant change in allele frequency between the Mendoza and Champagne populations of grapevine‐infecting B. cinerea is recorded. The large difference that was observed in the allele frequency of the molecular marker IGS/HaeIII is remarkable. More than 30% of the Mendoza population shared alleles that were recorded for the first time in the present study (alleles 5 and 6), and another 60% of the population shared an allele that was reported previously as exclusive for that population (private allele).7 This observation was confirmed by the F-statistics analysis, which gave a highly significant Fst value of 0.3332. This was much higher than that reported previously by Muñoz et al.18 between a Chilean population and the same Champagne population (Fst=0.109). It should be noted that Mendoza is close to the viticulture regions that were studied by Muñoz et al.18 (approximately 250km away) but it is isolated geographically from these regions by the Andes Mountains. Therefore, these areas act as completely isolated regions. The Fst values between the Champagne subpopulations were low as was observed for other comparisons of sites within a viticulture region.9,11 It should be noted that the latter data were obtained using microsatellite molecular markers, and not PCR-RFLP markers, which were used in the present study.
This study represents the first report of the genetic characterization of a B. cinerea population from Argentine vineyards. The information on genetic diversity that was obtained and the population differentiation data suggest the need of a further study, in order to establish the biological impact of these findings (e.g., virulence of the pathotypes and fungicide resistance).
Author's disclosure statementAuthors have nothing to declare. Authors have no conflict of interests.
This work was funded by the Instituto Nacional de Tecnología Agropecuaria (INTA), through its Project PNFRU 2185. The authors thank Ms. Fernanda Arias (EEA Mendoza INTA) and Dr. Evelyn Silva (Fundación Ciencia para la Vida, Santiago de Chile) for providing some samples used in this study. In addition, we wish to thank Dr. Tatiana Giraud (University of Paris Sud XI) for providing information about the Champagne population.