Fungi of the genus Paracoccidioides are the etiological agents of paracoccidioidomycosis, a highly prevalent mycosis in Latin America. Infection in humans occurs by the inhalation of conidia, which later revert to the form of yeast. In this context, macrophages are positioned as an important line of defense, assisting in the recognition and presentation of antigens, as well as producing reactive oxygen species that inhibit fungal spreading.
AimsThe objective of this study was to identify differentially expressed proteins during the interaction between Paracoccidioides lutzii Pb01 strain and human U937 monocytes.
MethodsTwo-dimensional electrophoresis, combined with mass spectrometry, was used to evaluate the differential proteomic profiles of the fungus P. lutzii (Pb01) interacting with U937 monocytes.
ResultsIt was possible to identify 25 proteins differentially expressed by Pb01 alone and after interacting with U937 monocytes. Most of these proteins are directly associated with fungal metabolism for energy generation, such as glyceraldehyde-3-phosphate dehydrogenase, and intracellular adaptation to monocytes. Antioxidant proteins involved in the response to oxidative stress, such as peroxiredoxin, cytochrome, and peroxidase, were expressed in greater quantity in the interaction with monocytes, suggesting their association with survival mechanisms inside phagocytic cells. We also identified 12 proteins differentially expressed in monocytes before and after the interaction with the fungus; proteins involved in the reorganization of the cytoskeleton, such as vimentin, and proteins involved in the response to oxidative stress, such as glioxalase 1, were identified.
ConclusionsThe results of this proteomic study of a P. lutzii isolate are novel, mimicking in vitro what occurs in human infections. In addition, the proteins identified may aid to understand fungal–monocyte interactions and the pathogenesis of paracoccidioidomycosis.
Los hongos del género Paracoccidioides son responsables de la paracoccidioidomicosis, una micosis muy prevalente en América Latina. La infección en el ser humano se produce por la inhalación de conidios, que después se transforman a la fase levaduriforme. En este contexto, los macrófagos asumen una importante línea de defensa, ayudando en el reconocimiento y presentación de antígenos, así como en la producción de especies reactivas del oxígeno que actúan para inhibir la proliferación de los hongos.
ObjetivosEl objetivo de este estudio fue identificar proteínas expresadas diferencialmente durante la interacción de la levadura Paraccocidioides lutzii Pb01 con macrófagos U937 humanos.
MétodosSe utilizó electroforesis bidimensional, conjuntamente con espectrometría de masas, para evaluar los perfiles proteómicos diferenciales del hongo P. lutzii (Pb01) en interacción con macrófagos U937.
ResultadosFue posible identificar 25 proteínas expresadas diferencialmente en Pb01 de manera aislada y tras la interacción con macrófagos U937. La mayoría de estas proteínas están directamente asociadas con el metabolismo de los hongos para la generación de energía, como la gliceraldehído-3-fosfato deshidrogenasa, y para la adaptación intracelular a los macrófagos. Las proteínas antioxidantes involucradas en la respuesta al estrés oxidativo, como la peroxirredoxina, el citocromo y la peroxidasa, se expresaron en mayor cantidad en la interacción con macrófagos, lo que sugiere su asociación con los mecanismos de supervivencia dentro de las células fagocíticas. Se identificaron 12 proteínas expresadas diferencialmente en macrófagos antes y después de la interacción con el hongo; se identificaron proteínas involucradas en la reorganización del citoesqueleto, como la vimentina, y proteínas involucradas en la respuesta al estrés oxidativo, como la glioxalasa 1.
ConclusionesLos resultados de este estudio proteómico de un aislamiento de P. lutzii son novedosos, y reflejan invitro lo que ocurre en las infecciones humanas. Además, las proteínas identificadas pueden ayudar a comprender las interacciones resultantes entre los hongos y los macrófagos y contribuir a la comprensión de la patogenia de la paracoccidioidomicosis.
Fungi in the genus Paracoccidioides are the etiological agents of paracoccidioidomycosis (PCM), one of the most prevalent systemic mycosis in Latin America, which affects individuals at a productive age and can lead to incapacitation, sequelae and, even, death.26,45 The fungus presents thermal dimorphism, growing as mycelia at temperatures below 28°C and as yeast in the host at 37°C.6,44,45 The genus Paracoccidioides comprises two species: Paracoccidioidesbrasiliensis, which represents a complex of four phylogenetically distinct species (S1, PS2, PS3, and PS4), and the species Paracoccidioideslutzii, which includes the isolate Pb01.10,30,31,47–49 This classification was recently refined following an analysis of genetic divergence to include species in the brasiliensis complex: PS2 (Paracoccidioidesamericana), PS3 (Paracoccidioidesrestrepiensis), and PS4 (Paracoccidioidesvenezuelensis).51
Fungus–host interactions trigger different cellular and molecular mechanisms.20 Once Paracoccidioides is inside a host cell, immunity mediated by cells in the innate and adaptive immune systems is activated.3,6,32 Macrophages and dendritic cells recognize and present fungal antigens to T lymphocytes, which produce Th1 cytokines such as tumor necrosis factor (TNF-α) and interferon gamma (IFN-γ), that activate macrophages to kill or inhibit the growth of the fungus.8 However, in some situations, Paracoccidioides can remain viable within macrophages by activating adaptive and defense mechanisms against conditions in the hostile environment.4,7 Patients with PCM have impaired cellular immunity with decreased levels of Th1 (IL-2, IFN-γ, and IL-12) cytokines and increased synthesis of Th2-standard cytokines (IL-4, IL-5, and IL-10).4
Protein profile analyses have been described for Paracoccidioides12,40,42 and human macrophages.34 The proteomic profiles of P. lutzii Pb01 have been elucidated in its transition from the mycelial to the pathogenic yeast phase42: differential expression of P. lutzii Pb01 proteins when compared to other species of Paracoccidioides in groups S1, PS2, and PS3,40 and during iron depletion,38 oxidative stress,9,23 and absence of carbon sources27; in interactions with murine macrophages,39 and membrane proteins in zinc deprivation conditions17; and differential metabolism of P. brasiliensis Pb03, Pb339, and PbEPM83 in a two-carbon substrate (sodium acetate).1 However, protein profile studies on the interactions between this pathogen and human macrophages have not yet been published and may constitute important information for an evaluation of PCM. These studies would help to elucidate the pathogenicity, virulence, and adaptive mechanisms in PCM, and to evaluate the ability of P. lutzii to survive stress conditions in the host. Thus, the objective of this study was to identify differentially expressed proteins during the interaction between P. lutzii Pb01 yeast and human U937 monocytes.
Material and methodsCulture of P. lutzii – Pb01The fungus Pb01 (kindly provided by Prof. Carlos Palleschi Taborda, University of São Paulo) was grown on Fava-Neto agar (1.3% peptone, 0.5% yeast extract, 0.5% meat extract, 4% glucose, 0.5% sodium chloride, and 1.3% agar, pH 7.2) for 15 days in an incubator (Bunker NI1521, São Paulo, Brasil) at 37°C.
Cell culture of U937 human monocytesU937 monocytes (Rio de Janeiro Cell Bank [BCRJ], Brazil) were maintained in 75cm2 culture bottles (Uniscience, Osasco, São Paulo, Brazil) with 25ml of RPMI (Sigma) medium plus 10% fetal bovine serum (FBS) (Gibco) and 1% penicillin-streptomycin (10μg/ml) (Sigma) at 37°C in a humidified atmosphere with 5% CO2 for 72h, and a final concentration of 1×106cells/ml.
Interactions between U937 monocytes and P. lutzii – Pb01U937 monocytes at a concentration of 1×106cells/ml and fungal strain Pb01 at 2×106cells/ml were maintained in RPMI medium with 10% FBS and 1% penicillin-streptomycin (10μg/m) in incubator (CCL-050B-9, Uniscience, Osasco, São Paulo, Brazil) at 37°C, 95% of relative humidity and 5% CO2 for 72h. In order to examine the P. lutzii Pb01 strain and the monocyte cell line U937 at 72h of interaction (the time that showed the most activity, data not shown), the production of IL-10, IL-12, TNF-α, and IFN-γ cytokines in supernatants was analyzed by enzyme-linked immunosorbent assay (ELISA). Furtheremore, using flow cytometry we analized peripheral blood mononuclear cells (PBMC), 1×106cells/ml, at 0, 24, and 72h before and after the interaction with fungal cells Pb01 at a concentration of 2×106cells/ml. In this sample we measured the CD4 and CD8 molecules on the surfaces of lymphocytes, and CD14 and CD86 on the surfaces of monocytes. The data obtained were used for an internal evaluation of the cell line screening after the interaction with P. lutzii.
Extraction of proteinsProteins were extracted using the method of Fonseca et al. (2001)19 and Pirovani et al. (2008)41 with some modifications. The RPMI broths with U937 monocytes alone, P. lutzii Pb01 strain and both were washed three times with 1×PBS by centrifugation at 10,000×g for 10min. The cells were macerated in liquid nitrogen (N2), supplemented with protease inhibitor cocktail (Sigma), and sonicated (amplitude of 70%, three pulses of 10s each, with intervals of 60s) on ice. To precipitate the proteins, an equivalent amount of 20% trichloroacetic acid was added; the solution was kept at −20°C for 60min and then centrifuged at 16,100×g for 10min. The supernatant was discarded, and the cell pellet was washed with 500μl of absolute acetone, sonicated (70% amplitude, three pulses of 7s each at 21-s intervals) on ice, and centrifuged for 10min at 16,100×g. Subsequently, the pellet was washed two times with 0.1M ammonium acetate and two times with 80% acetone, dried under vacuum, and resuspended in rehydration buffer (7moll−1 urea, 2moll−1 thiourea, 2% CHAPS, and 0.002% bromophenol blue) to solubilize the proteins. The proteins were quantitated using a 2D Quant kit (GE Healthcare) according to the manufacturer's instructions. Samples were stored at −20°C until use.
1D and 2D electrophoresisFor 1D electrophoresis, 30μg of each protein extract (U937 monocyte, Pb01 strain, and U937-Pb01) were separated on 12.5% polyacrylamide gels using sodium dodecyl sulfate-polyacrylamide electrophoresis (SDS-PAGE). For 2D electrophoresis, 350μg of the protein extracts were used. Protein samples were put onto strips of 13cm in length, with an immobilized pH gradient of 3–10 (Amersham Biosciences, Immobiline Dry-Strip). The strips were subjected to isoelectric focusing under the following conditions: 1h at 500Vh, 1h 40min at 1000Vh, 2h 30min at 2200Vh, and 4h 35min at 8000Vh. Strips were treated with 7ml of equilibration buffer (50mM Tris, 6M urea, 30% glycerol, and 2% SDS) containing 1% w/v dithiothreitol for 15min under slow stirring, and with a buffer containing 2.5% iodoacetamide for another 15min. The strips were then washed with running buffer (Tris 0.025mol/l, pH 8.3, 0.19mol/l glycine, 0.1% SDS) for 15min. For the second dimension, strips were transferred to a 12.5% SDS-PAGE gel in a Ruby SE600 (GE Healthcare) vertical electrophoresis system, and an initial electric current of 15mA/gel was applied for 15min, followed by 50mA/gel for 3h and 30min. Gels were prepared in triplicate for each sample. The polyacrylamide gels were fixed (40% ethanol and 10% acetic acid) for 1h and stained with Coomassie blue.
Protein visualization and image analysisThe images of the gels were scanned using an image scanner (GE Healthcare) with the LabScanner software (Amersham Biosciense), and analyzed using Image Master 2D Platinum 7.0 (GE Healthcare) software for the identification and quantification (based on the area, volume and intensity of the spots) of the proteins from Pb01 and the interaction between Pb01 and monocytes. Gels in triplicate were used to determine the relative accumulation of proteins from Pb01 extract and Pb01 together with U937 monocytes extract. Common spots that had their expression differentially increased or decreased based on the ratio of the percentage of normalized volume (% vol) by a factor 1.5 times (fold) were considered of interest. An ANOVA test (analysis of variance) was used, and differences were considered statistically significant when p<0.05.
Extraction of peptidesThe spots selected were manually excised directly from the replicate colloidal Coomassie Blue-SDS PAGE gels and destained in 50% (v/v) acetonitrile (Fisher Scientific)/25mM NH4HCO3 (Sigma), pH 8.0, until clearing of blue stain. The gel fragments were dehydrated in 100% acetonitrile and completely dried in a SpeedVac (Eppendorf). In-gel trypsin digestion was performed using 20μg/ml of trypsin Gold (Promega, Madison, WI) in 25mM NH4HCO3, pH 8.0, at 37°C for 24h. Tryptic peptides were extracted using 5% (v/v) formic acid (Merck)/50% (v/v) acetonitrile. Peptides were then concentrated in a SpeedVac to approximately 10μl and stored at −20°C. Subsequently, 40μl 25mM NH4HCO3, pH 8.0, were added to the peptides and then concentrated in a SpeedVac to approximately 10μl for mass spectrometry analysis at the Laboratory of Proteomics, Center of Biotechnology and Genetics – Universidade Estadual de Santa Cruz – UESC, Ilheús (Bahia, Brazil).
Mass spectrometryThe peptides obtained were resolved by reverse phase chromatography on a nanoAquity UPLC (Waters, Milford, MA, USA) using two C18 columns (the first was a trapping column of 5μm and 180μm×20mm, and the second was 1.7μm and 100μm×100mm) under a flow of 0.6μl/min in a 50-min run. The peptides were separated on an acetonitrile gradient, transferred to a mass spectrometer (Micromass Q-TOFmicro, Waters), and ionized in a 3000-V capillary. The peptides were fragmented in positive mode with a minimum relative intensity of 10 counts. The three most intense ions were analyzed in 1-s scans, with collision energy ranging from 20 to 95eV according to the mass/charge (m/z) ratio of the peptides.
Identification and characterization of proteinsThe mass spectra obtained were analyzed using MASCOT server online software (Matrix Science: http://www.matrixsciesce.com) and compared in the non-redundant National Biotechnology Information Center (NCBInr) database. The search parameters used in MASCOT were the following: tryptic digestion with a lost cleavage site, methionine oxidation and cysteine carboxamidomethylation as variable, and tolerance of peptide masses of 0.3Da and mass tolerance of 0.1-Da fragments as fixed modifications. Based on the MASCOT analysis, only those peptides with a statistical significance of p<0.05 were accepted and used to validate the proteins. Functional characterization of the identified proteins was performed by means of biological process annotations using BLAST2GO software (http://www.blast2go.com).
The differential expression profiles of the identified proteins were analyzed by hierarchical clustering. The R program with Heatmap2 package was used to construct a heatmap. The expression values for each spot (% of volume), determined using the ImageMaster 2D Platinum 7.0 (GE Healthcare) gel image analysis program, were transformed into Z-scores (data not shown).
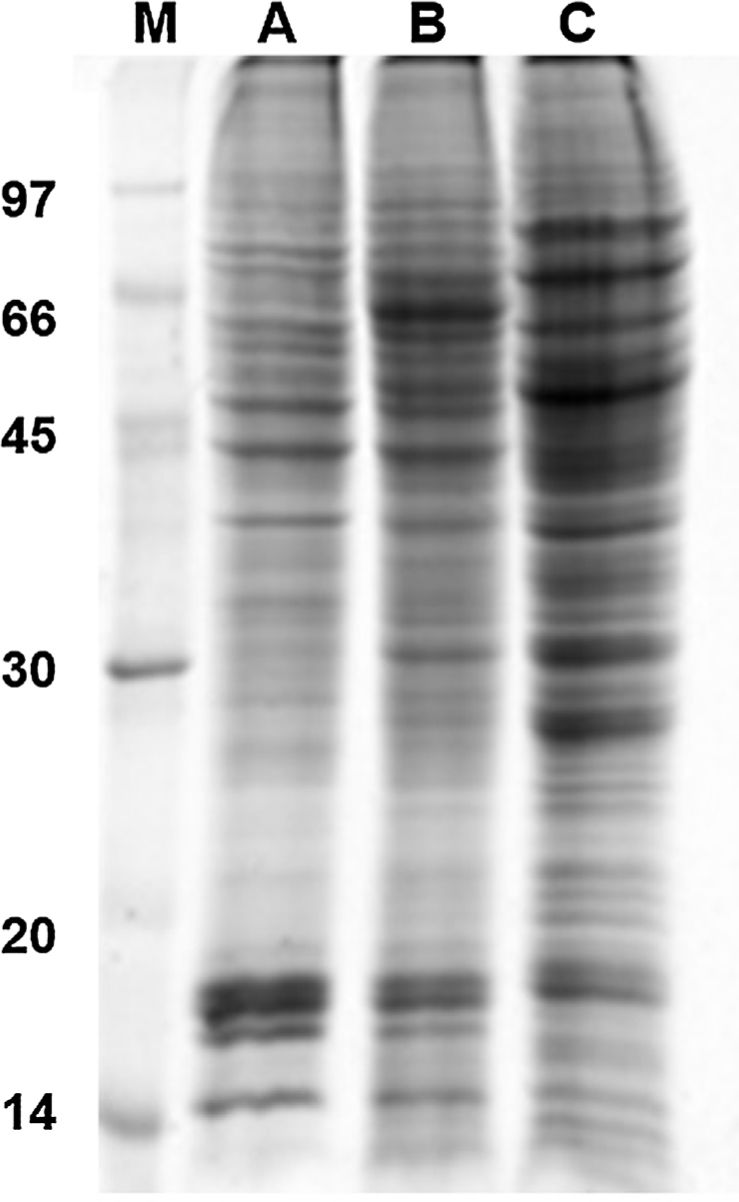
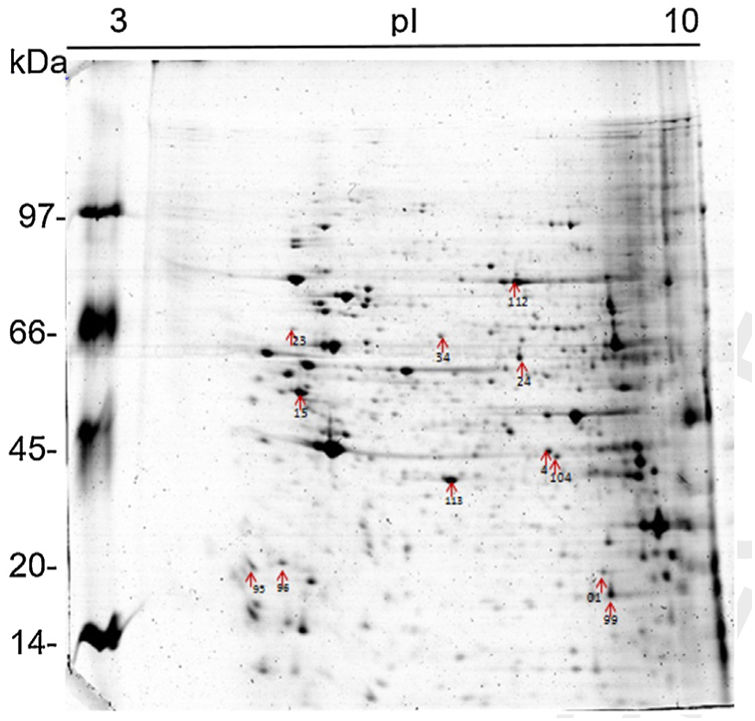
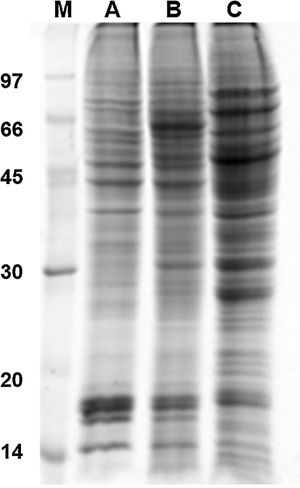
Results and discussion1D and 2D protein profilesTo analyze fungus–monocyte interactions, the U937 cells were maintained alone and in the presence of Pb01 for 72h. The 1D gel of U937 monocytes proteins alone or with Pb01 showed bands with molecular weights ranging from 14 to 97kDa. In relation to the molecular weight of Pb01 proteins before and after interaction, great variability was observed, with a predominance of bands with high molecular weights (Fig. 1).
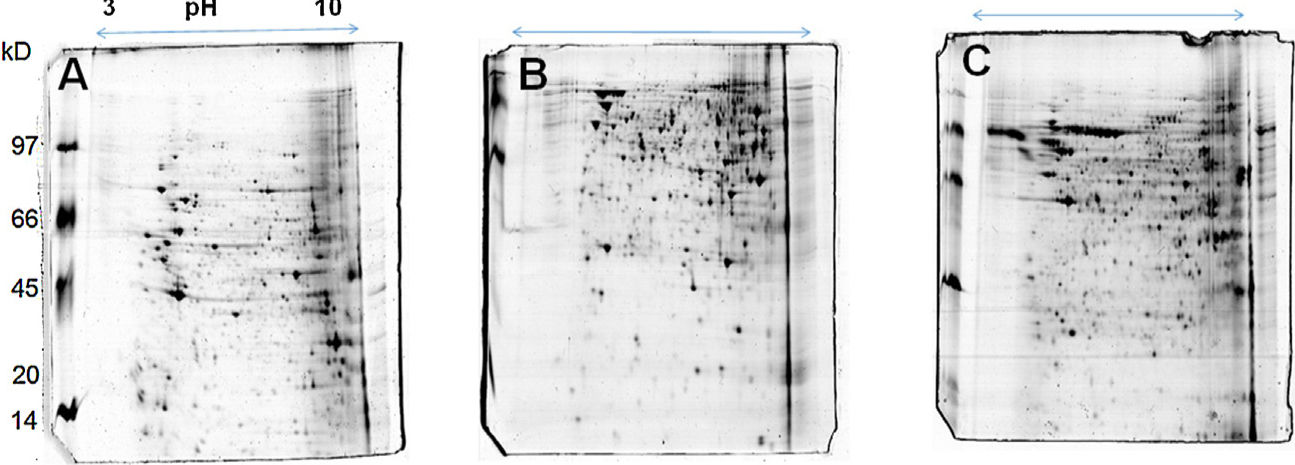
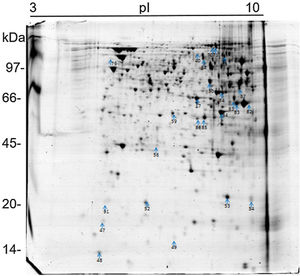
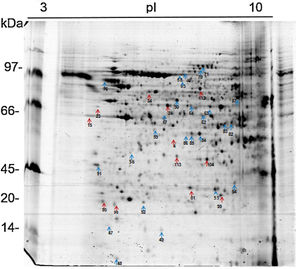
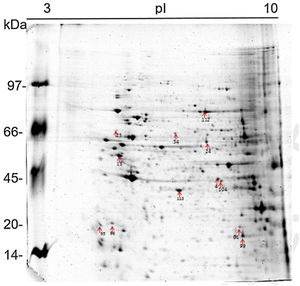
The 2D gel revealed a total of 263 spots with U937 alone, 282 spots with Pb01, and 373 with U937+Pb01. Proteins were visualized at all isoelectric points (pI) and molecular weights, with a predominance of proteins with molecular weights above 30kDa, corroborating the gel bands obtained by 1D electrophoresis (Fig. 2).
In this study, spots from the three treatments were observed from 8 to 122kDa and 3 to 10pI. The spots of Pb01 presented values from 9 to 120kDa and 4 to 10pI. In the U937 monocytes spots, the pI was between 4 and 10, and molecular weight was between 20 and 40kDa. Similarly to the results of this study, but using alveolar monocytes, Minafra et al.33 observed differentially expressed proteins with pIs from 4.96 to 8.37 and molecular weights from 22 to 56kDa. Pigosso et al.,40 also studying Pb01, observed less molecular weight variation, between 11 and 84kDa, and pIs from 3 to 11.
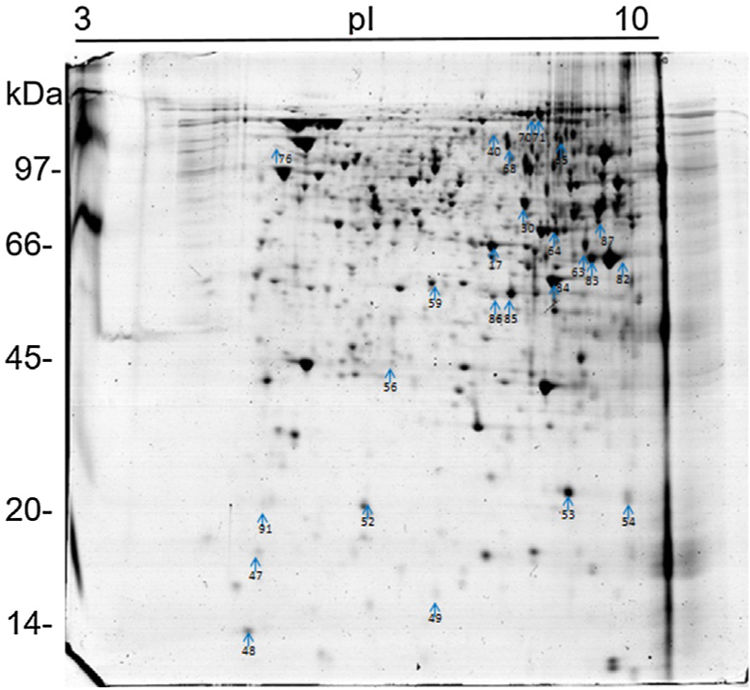
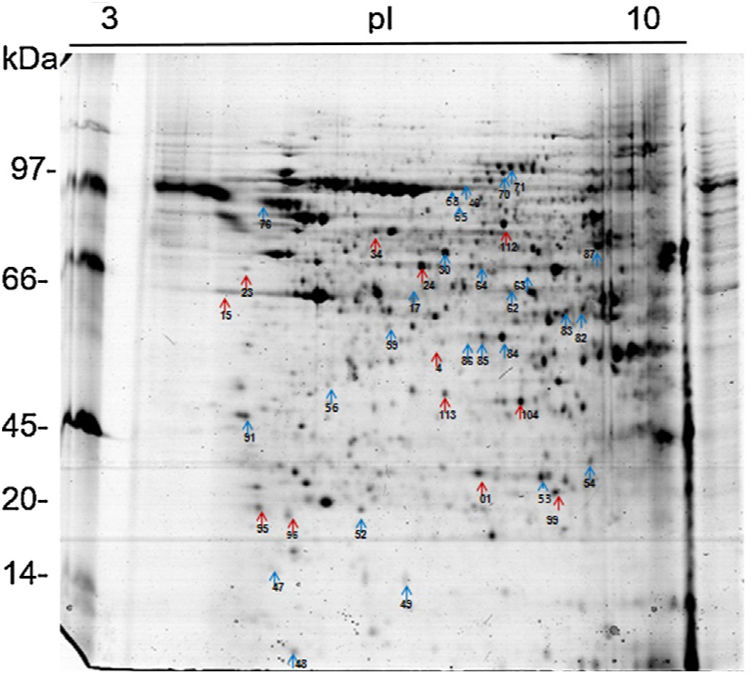
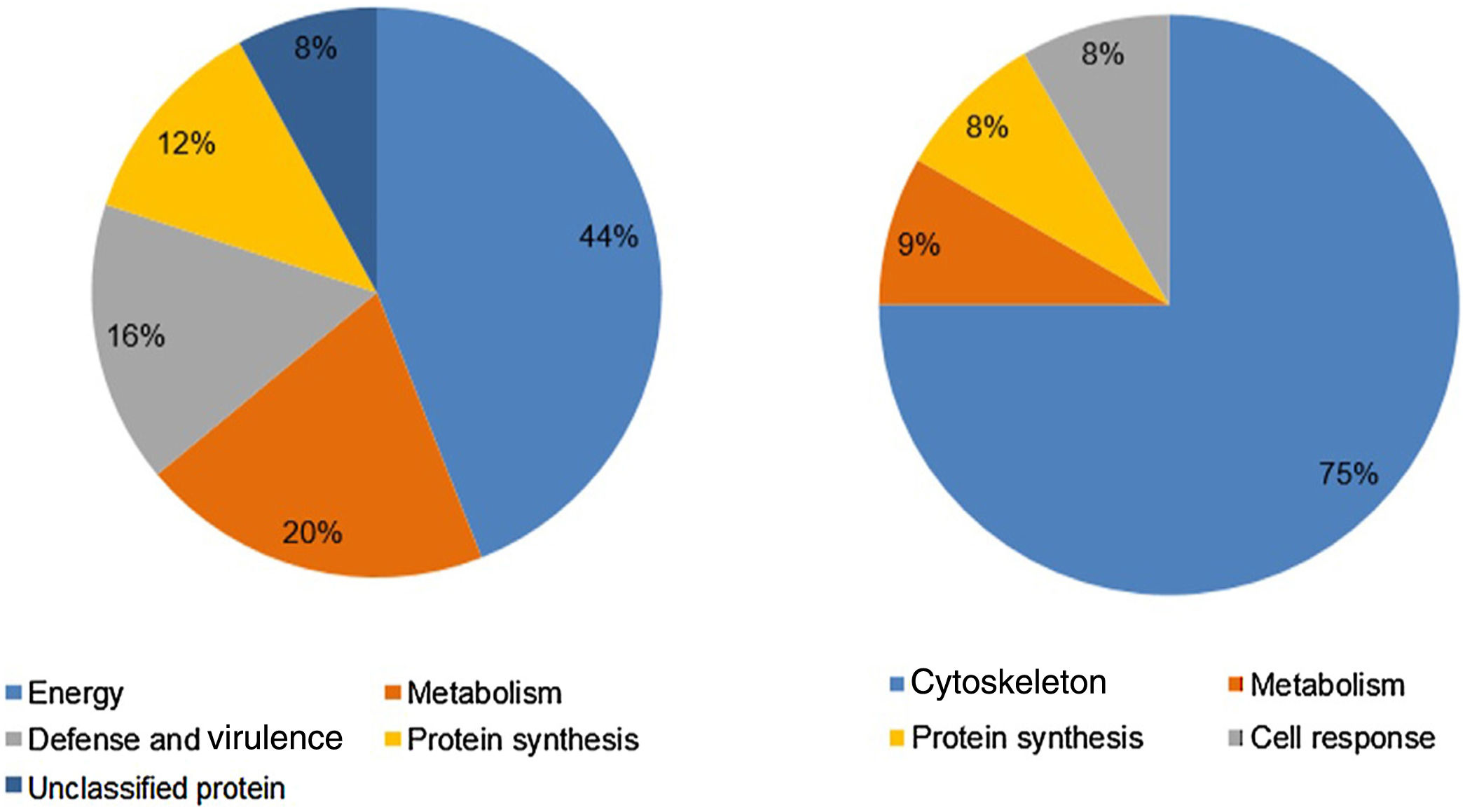
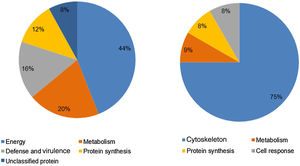
Analysis, identification, and classification of proteins differentially expressed in Pb01 alone and with monocytesThe analysis revealed a total of 25 spots of proteins differentially expressed by Pb01 and Pb01+monocytes, and 12 spots differentially expressed by monocytes and Pb01+monocytes, that were identified by mass spectrometry (Figs. 3–5) (Tables 1 and 2). The proteins were classified by the Blast2GO software in relation to the biological processes in which they can participate. The proteins from Pb01 were distributed in the following functional categories: energy and metabolism, defense and virulence, and synthesis of proteins and hypothetical proteins (Fig. 6).
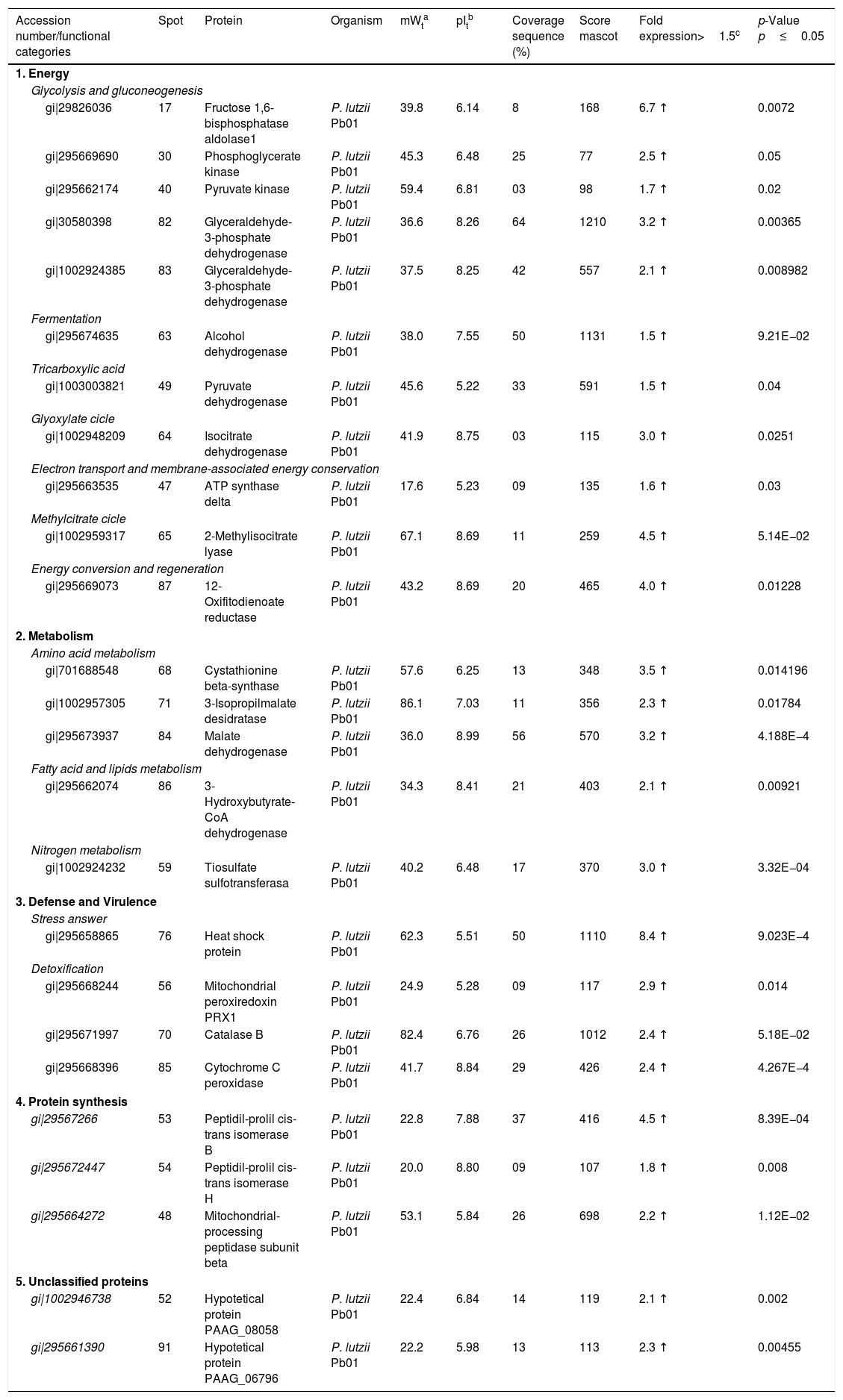
Identification of differentially expressed proteins in Pb01 after interaction with U937 macrophages.
| Accession number/functional categories | Spot | Protein | Organism | mWta | pItb | Coverage sequence (%) | Score mascot | Fold expression>1.5c | p-Value p≤0.05 |
|---|---|---|---|---|---|---|---|---|---|
| 1. Energy | |||||||||
| Glycolysis and gluconeogenesis | |||||||||
| gi|29826036 | 17 | Fructose 1,6-bisphosphatase aldolase1 | P. lutzii Pb01 | 39.8 | 6.14 | 8 | 168 | 6.7 ↑ | 0.0072 |
| gi|295669690 | 30 | Phosphoglycerate kinase | P. lutzii Pb01 | 45.3 | 6.48 | 25 | 77 | 2.5 ↑ | 0.05 |
| gi|295662174 | 40 | Pyruvate kinase | P. lutzii Pb01 | 59.4 | 6.81 | 03 | 98 | 1.7 ↑ | 0.02 |
| gi|30580398 | 82 | Glyceraldehyde-3-phosphate dehydrogenase | P. lutzii Pb01 | 36.6 | 8.26 | 64 | 1210 | 3.2 ↑ | 0.00365 |
| gi|1002924385 | 83 | Glyceraldehyde-3-phosphate dehydrogenase | P. lutzii Pb01 | 37.5 | 8.25 | 42 | 557 | 2.1 ↑ | 0.008982 |
| Fermentation | |||||||||
| gi|295674635 | 63 | Alcohol dehydrogenase | P. lutzii Pb01 | 38.0 | 7.55 | 50 | 1131 | 1.5 ↑ | 9.21E−02 |
| Tricarboxylic acid | |||||||||
| gi|1003003821 | 49 | Pyruvate dehydrogenase | P. lutzii Pb01 | 45.6 | 5.22 | 33 | 591 | 1.5 ↑ | 0.04 |
| Glyoxylate cicle | |||||||||
| gi|1002948209 | 64 | Isocitrate dehydrogenase | P. lutzii Pb01 | 41.9 | 8.75 | 03 | 115 | 3.0 ↑ | 0.0251 |
| Electron transport and membrane-associated energy conservation | |||||||||
| gi|295663535 | 47 | ATP synthase delta | P. lutzii Pb01 | 17.6 | 5.23 | 09 | 135 | 1.6 ↑ | 0.03 |
| Methylcitrate cicle | |||||||||
| gi|1002959317 | 65 | 2-Methylisocitrate lyase | P. lutzii Pb01 | 67.1 | 8.69 | 11 | 259 | 4.5 ↑ | 5.14E−02 |
| Energy conversion and regeneration | |||||||||
| gi|295669073 | 87 | 12-Oxifitodienoate reductase | P. lutzii Pb01 | 43.2 | 8.69 | 20 | 465 | 4.0 ↑ | 0.01228 |
| 2. Metabolism | |||||||||
| Amino acid metabolism | |||||||||
| gi|701688548 | 68 | Cystathionine beta-synthase | P. lutzii Pb01 | 57.6 | 6.25 | 13 | 348 | 3.5 ↑ | 0.014196 |
| gi|1002957305 | 71 | 3-Isopropilmalate desidratase | P. lutzii Pb01 | 86.1 | 7.03 | 11 | 356 | 2.3 ↑ | 0.01784 |
| gi|295673937 | 84 | Malate dehydrogenase | P. lutzii Pb01 | 36.0 | 8.99 | 56 | 570 | 3.2 ↑ | 4.188E−4 |
| Fatty acid and lipids metabolism | |||||||||
| gi|295662074 | 86 | 3-Hydroxybutyrate-CoA dehydrogenase | P. lutzii Pb01 | 34.3 | 8.41 | 21 | 403 | 2.1 ↑ | 0.00921 |
| Nitrogen metabolism | |||||||||
| gi|1002924232 | 59 | Tiosulfate sulfotransferasa | P. lutzii Pb01 | 40.2 | 6.48 | 17 | 370 | 3.0 ↑ | 3.32E−04 |
| 3. Defense and Virulence | |||||||||
| Stress answer | |||||||||
| gi|295658865 | 76 | Heat shock protein | P. lutzii Pb01 | 62.3 | 5.51 | 50 | 1110 | 8.4 ↑ | 9.023E−4 |
| Detoxification | |||||||||
| gi|295668244 | 56 | Mitochondrial peroxiredoxin PRX1 | P. lutzii Pb01 | 24.9 | 5.28 | 09 | 117 | 2.9 ↑ | 0.014 |
| gi|295671997 | 70 | Catalase B | P. lutzii Pb01 | 82.4 | 6.76 | 26 | 1012 | 2.4 ↑ | 5.18E−02 |
| gi|295668396 | 85 | Cytochrome C peroxidase | P. lutzii Pb01 | 41.7 | 8.84 | 29 | 426 | 2.4 ↑ | 4.267E−4 |
| 4. Protein synthesis | |||||||||
| gi|29567266 | 53 | Peptidil-prolil cis-trans isomerase B | P. lutzii Pb01 | 22.8 | 7.88 | 37 | 416 | 4.5 ↑ | 8.39E−04 |
| gi|295672447 | 54 | Peptidil-prolil cis-trans isomerase H | P. lutzii Pb01 | 20.0 | 8.80 | 09 | 107 | 1.8 ↑ | 0.008 |
| gi|295664272 | 48 | Mitochondrial-processing peptidase subunit beta | P. lutzii Pb01 | 53.1 | 5.84 | 26 | 698 | 2.2 ↑ | 1.12E−02 |
| 5. Unclassified proteins | |||||||||
| gi|1002946738 | 52 | Hypotetical protein PAAG_08058 | P. lutzii Pb01 | 22.4 | 6.84 | 14 | 119 | 2.1 ↑ | 0.002 |
| gi|295661390 | 91 | Hypotetical protein PAAG_06796 | P. lutzii Pb01 | 22.2 | 5.98 | 13 | 113 | 2.3 ↑ | 0.00455 |
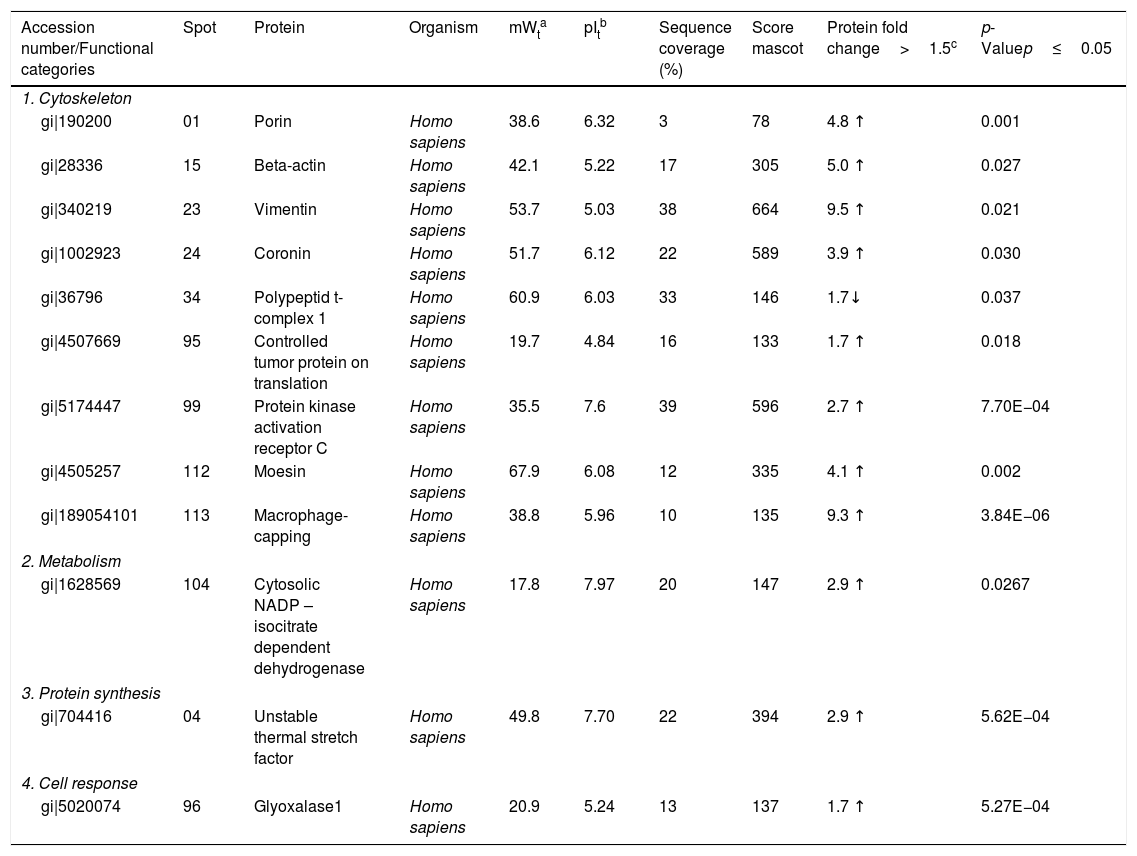
Identification of differentially expressed proteins in U937 macrophages after interaction with Pb01.
| Accession number/Functional categories | Spot | Protein | Organism | mWta | pItb | Sequence coverage (%) | Score mascot | Protein fold change>1.5c | p-Valuep≤0.05 |
|---|---|---|---|---|---|---|---|---|---|
| 1. Cytoskeleton | |||||||||
| gi|190200 | 01 | Porin | Homo sapiens | 38.6 | 6.32 | 3 | 78 | 4.8 ↑ | 0.001 |
| gi|28336 | 15 | Beta-actin | Homo sapiens | 42.1 | 5.22 | 17 | 305 | 5.0 ↑ | 0.027 |
| gi|340219 | 23 | Vimentin | Homo sapiens | 53.7 | 5.03 | 38 | 664 | 9.5 ↑ | 0.021 |
| gi|1002923 | 24 | Coronin | Homo sapiens | 51.7 | 6.12 | 22 | 589 | 3.9 ↑ | 0.030 |
| gi|36796 | 34 | Polypeptid t-complex 1 | Homo sapiens | 60.9 | 6.03 | 33 | 146 | 1.7↓ | 0.037 |
| gi|4507669 | 95 | Controlled tumor protein on translation | Homo sapiens | 19.7 | 4.84 | 16 | 133 | 1.7 ↑ | 0.018 |
| gi|5174447 | 99 | Protein kinase activation receptor C | Homo sapiens | 35.5 | 7.6 | 39 | 596 | 2.7 ↑ | 7.70E−04 |
| gi|4505257 | 112 | Moesin | Homo sapiens | 67.9 | 6.08 | 12 | 335 | 4.1 ↑ | 0.002 |
| gi|189054101 | 113 | Macrophage-capping | Homo sapiens | 38.8 | 5.96 | 10 | 135 | 9.3 ↑ | 3.84E−06 |
| 2. Metabolism | |||||||||
| gi|1628569 | 104 | Cytosolic NADP – isocitrate dependent dehydrogenase | Homo sapiens | 17.8 | 7.97 | 20 | 147 | 2.9 ↑ | 0.0267 |
| 3. Protein synthesis | |||||||||
| gi|704416 | 04 | Unstable thermal stretch factor | Homo sapiens | 49.8 | 7.70 | 22 | 394 | 2.9 ↑ | 5.62E−04 |
| 4. Cell response | |||||||||
| gi|5020074 | 96 | Glyoxalase1 | Homo sapiens | 20.9 | 5.24 | 13 | 137 | 1.7 ↑ | 5.27E−04 |
The main Pb01 proteins identified by proteomic analysis were related to energy metabolism. An increased expression of glycolysis and gluconeogenesis proteins, especially fructose 1,6 bisphosphate aldolase (FBA) (spot 17) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (spots 82 and 83), was observed when Pb01 interacted with monocytes. These proteins have already been identified in Paracoccidioides19; they are related to virulence processes such as fungus adhesion and interaction with host proteins,29 as well as immunogenicity, phagocytosis, and binding to human plasminogen for tissue invasion and fungus dissemination.11 GAPDH expression was previously described in P. brasiliensis under conditions that mimic the hematological pathway of fungal spread, and has been found also on the surface of Candida albicans cells, suggesting that this protein participates in the fungal infection process.2 The protein isocitrate dehydrogenase (spot 64), which participates in the glyoxylate cycle, was also highly expressed after the interaction of the fungus with monocytes. Concerning this protein, an increased gene expression was described in the species P. brasiliensis18 and in other fungi such as C. albicans,28Cryptococcus neoformans,43 and Penicillium marneffei,50 suggesting that this protein is associated with energy synthesis in intracellular environments, for example during phagocytosis. Similarly, enzyme 2-methylisocitrate (spot 65) also showed increased expression after Pb01 interaction with monocytes, revealing that P. lutzii uses this mechanism to generate energy via the methylcitrate cycle.10 Also in relation to metabolism and energy, a higher expression of alcohol dehydrogenase enzyme (spot 63) was observed in Pb01 after the interaction with monocytes, suggesting that P. lutzii can use anaerobic glycolysis pathways to generate energy in monocyte microenvironments.39,40 The same enzyme was described in an experimental model of Aspergillus fumigatus infection, and its presence was associated with hypoxic conditions and fungal pathogenesis.22,52–54
Other important metabolic adaptations of P. lutzii in response to phagocytosis were increased expression of proteins involved in the metabolism of amino acids, such as cystathione beta synthase (spot 68), 3-isopropylmalate dehydrogenase (spot 71) and malate dehydrogenase (spot 84); in the metabolism of fatty acids, such as 3-hydroxybutyrate-CoA dehydrogenase (spot 86); and in the metabolism of nitrogen, such as thiosulfate sulfotransferase (spot 59). This energy group protein profile suggests an adaptive response of P. lutzii to survive inside the inhospitable environment of monocytes, where nutrient concentrations may be relatively low.14,46
Higher levels of the enzymes peroxiredoxin PRX1, catalase, and cytochrome C peroxidase antioxidant (spots 56, 70, and 85, respectively) were also observed in Pb01 after the interaction with U937. These enzymes are involved in detoxification by reacting with certain molecules such as reactive oxygen species (ROS), radical hydroxyl (OH), superoxide anion (O2), and hydrogen peroxide (H2O2), which are produced by macrophages during phagocytosis.8 In the fungus C. neoformans, these enzymes were associated with virulence, as well as survival of the fungus in a macrophage oxidative environment.34,35 The enzymes cytochrome C peroxidase and peroxiredoxin PRX1 play antioxidant roles in the response of P. brasiliensis to oxidative stress during the interaction with murine macrophages cells25,39,46; however, in P. lutzii, these enzymes were not previously identified, indicating a pioneering finding.
Also, in response to oxidative stress, heat shock protein HSP60 (spot 76) was detected in Pb01 after the interaction with monocytes. Paracoccidioides Hsp60 and Hsp70 proteins are important immunogenic proteins because they are fungal antigens recognized by antibodies in sera from patients with PCM.15 HSP60 expression has been found to contribute to changes in Histoplasma capsulatum cell walls that allow the fungus to survive under stress conditions such as thermal stress, infection, and pathogenesis.24 Increased levels of heat shock proteins have also been observed in Pb01 in murine infections, suggesting that these proteins are necessary for the survival of the fungus under the thermal conditions of the host and that they may play a role in the morphogenesis of P. lutzii.13,15,39
Analysis, identification, and classification of proteins differentially expressed in U937 alone and in the interaction with Pb01The proteins identified in U937 monocytes before and after their interaction with Pb01 were distributed in the functional categories of cytoskeleton, metabolism, and protein synthesis and cellular defense (Fig. 6). Most of the proteins identified by proteomic analysis in U937 monocytes after the interaction with Pb01 are associated with cellular cytoskeleton activity. Increased levels of expression, mainly of porins (spot 01) beta actin (spot 15), vimentin (spot 23), moesin (spot 112), and monocyte-capping proteins (spot 113), are related to the formation of pseudopodia during phagocytosis, which requires changes in cell morphology and cytoskeletal rearrangements.33 Vimentin is involved in the rearrangement of the cytoskeleton and is abundant in human monocytes.37 Some studies have demonstrated the role of this protein during phagocytosis in response to infectious agents.5,37
Monocytes-capping protein is related to the maturation of monocytes into macrophages and may play an important role in the activation of macrophages.16 The protein glioxalase 1 (spot 96) also showed increased expression in U937 monocytes after the interaction with Pb01. This protein is constitutively expressed in mammalian cells and is associated with detoxification activity; its function is essential for the cellular protection against glycation and the oxidative stress generated by infectious agent-macrophage interactions.36 The presence of antioxidant enzymes has been shown in a proteomic analysis of infected alveolar monocytes, suggesting that they play a role in microbicidal mechanisms, essential for the eradication of a pathogen.25 In murine macrophages, C. albicans antioxidant enzymes increased the host response required to destroy the pathogen and control candidiasis development.21
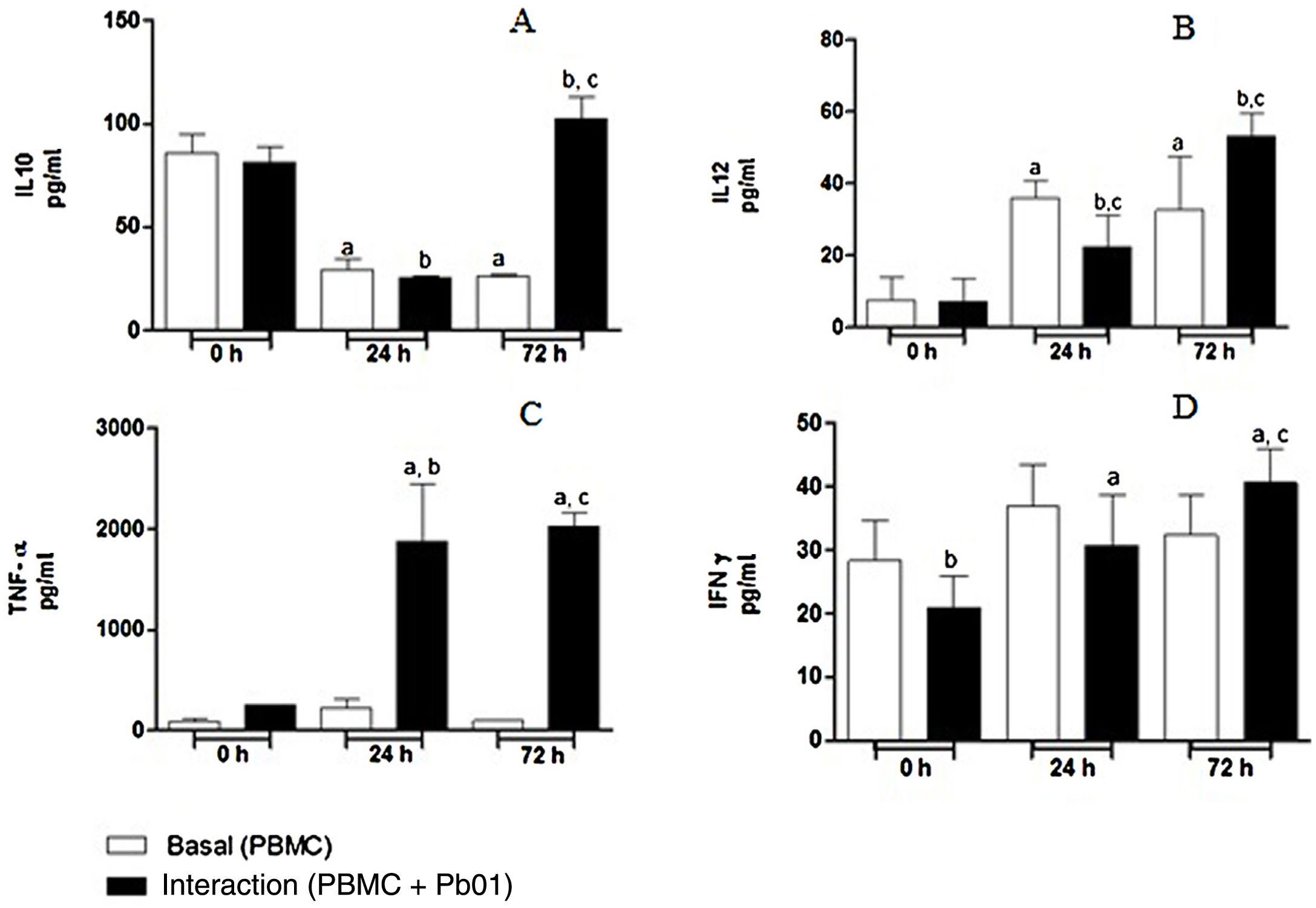
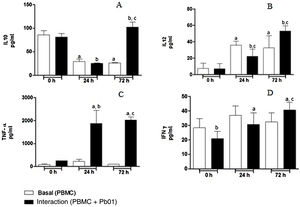
Kinetics of cytokine production by PBMC after the interaction with P. lutziiRegarding the production of IL-10, an anti-inflammatory cytokine, there was a statistically significant increase only after 72h of fungus-cell interaction compared to the baseline condition (control) in the same period. These levels were higher than those in the interaction at 0 and 24h. Likewise, the highest levels of IL-12, a pro-inflammatory cytokine, were also produced after 72h of interaction, and this increase was significantly greater when compared to the interactions at 0 and 24h. In the analysis of TNF-α, also a pro-inflammatory cytokine, a statistically significant production was observed both after 24 and 72h of interaction; under the stimulation of the fungus, the highest production also occurred after 72h. In relation to IFN-γ, a greater production occurred under antigenic stimulus after 72h of interaction, although higher levels of this cytokine were also detected at baseline and after 24h in cells maintained only in culture medium (Fig. 7).
Concentration (pg/ml) of cytokines in mononuclear cells of peripheral blood before and after sensitization with P. lutzii cells. A: IL-10; B: IL-12; C: TNF-α; D: IFN-γ (pg/ml), (A): a: significant difference when compared with the baseline (PBMC alone) at 0h; b: significant difference when compared with the interaction (PBMC+Pb01) at 0h; c: significant difference when compared with the baseline (PBMC alone) at 72h. (B): a: significant difference when compared with the baseline (PBMC alone) at 0h; b: significant difference when compared with the interaction (PBMC+Pb01) at 0h; c: significant difference when compared with the baseline (PBMC alone) at the same time. (C): a: significant difference when compared to the interaction (PBMC+Pb01) at 0h; b: significant difference when compared with the baseline (PBMC alone) at 24h; c: significant difference when compared with the baseline (PBMC alone) at 72h. (D): a: significant difference when compared to the interaction (PBMC+Pb01) at 0h; b: significant difference when compared with the baseline (PBMC alone) at 72h. Values are the average of the results of three assays+standard deviation.
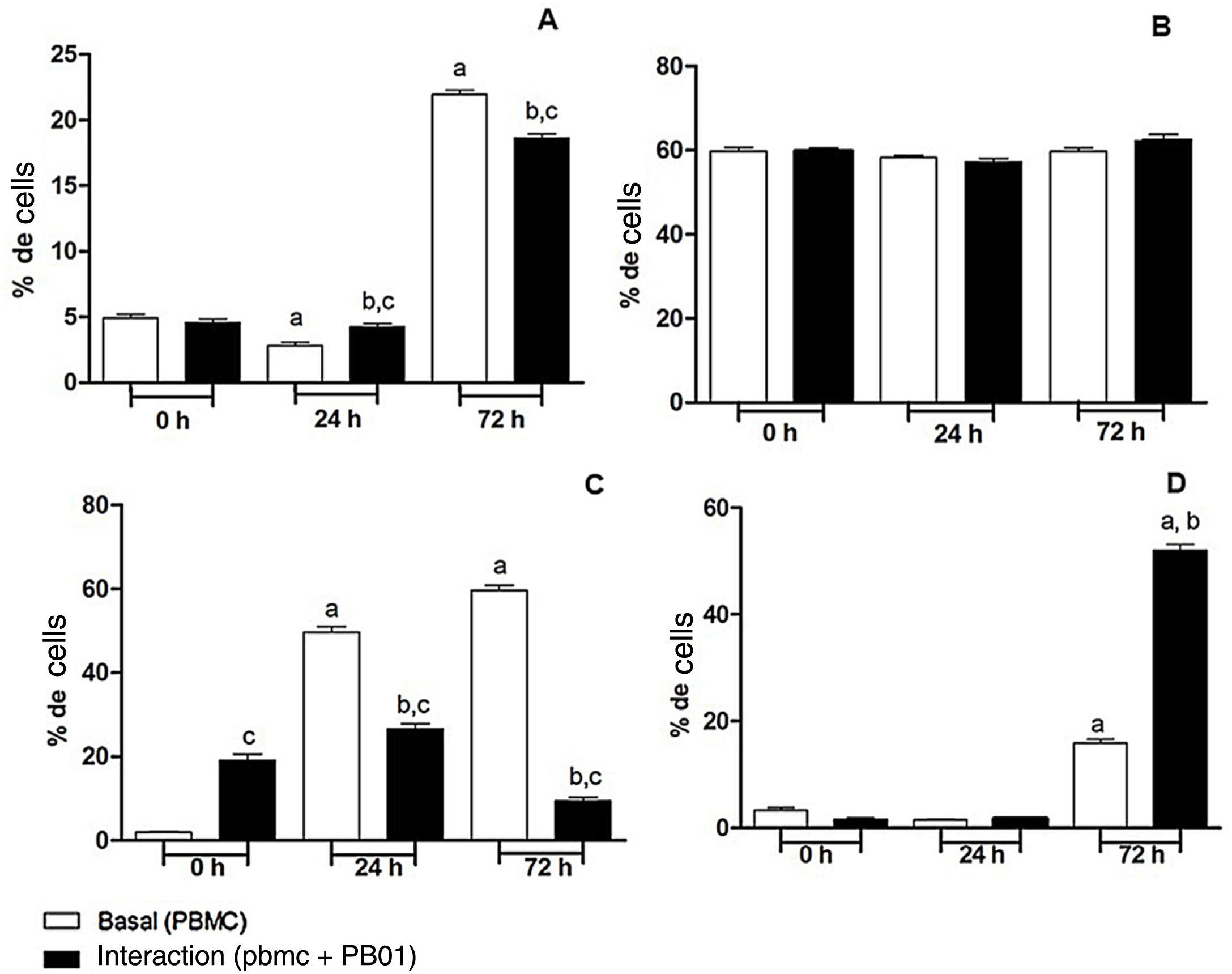
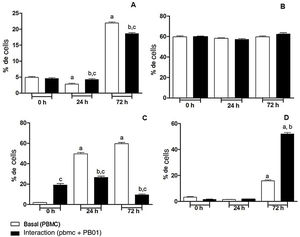
In the present study we also assessed the expression of the CD4 receptor (helper cells), CD8 (cytotoxic activity), CD14 (macrophage marker) and CD86 (macrophage activation) of mononuclear cells from peripheral blood (PBMC) at 0, 24 and 72h, before and after the interaction with cells of the fungus P. lutzii. It was observed that Pb01 in interaction with PBMC increased CD14 expression only after 72h and showed no change in the expression pattern compared to CD86 under stimulus with P. lutzii. In relation to CD4, the lymphocytes showed a reduction in the expression of this receptor after antigenic stimulation, regardless of the culture time (24 and 72h). The expression of CD8 was observed only after 72h of interaction with the fungus (Fig. 8).
Expression of surface molecules by peripheral blood mononuclear cells before and after sensitization with P. lutzii cells. A: CD14; B: CD86; C: CD4; D: CD8. (A): a: significant difference when compared with the baseline (PBMC alone) at 0h; b: significant difference when compared with the interaction (PBMC+Pb01) at 0h; c: significant difference when compared with the baseline (PBMC alone) at the same time. (B): there was no significant difference at 0, 24 and 72h. (C): a: significant difference when compared with the baseline (PBMC alone) at 0h; b: significant difference when compared with the interaction (PBMC+Pb01) at 0h; c: significant difference when compared with the baseline (PBMC alone). (D): a: significant difference when compared with the baseline (PBMC alone) at 0h; b: significant difference when compared with the interaction (PBMC+Pb01) at 0h. Values are the average of the results of three assays+standard deviation.
The results of the proteomic analysis of the fungus P. lutzii and human monocytes increase our knowledge of the molecular mechanisms involved in fungus-host interactions, suggesting that many Pb01 proteins are expressed after the interaction with monocytes to facilitate immune adaptation and escape.
Originality of the materialThe content of this article is original and has not been previously published nor sent or considered for any other publication, in whole or in any of its parts.
AuthorshipAll authors read and approved the manuscript and the authorship requirements were met.
Conflicts of interestThe authors declare that there are no conflicts of interest related to the publication of this article.