Phytophthora is the most important genus of the Oomycete plant pathogens. Nowadays, there are 117 described species in this genus, most of them being primary invaders of plant tissues. The different species are causal agents of diseases in a wide range of crops and plants in natural environments. In order to develop control strategies against Phytophthoraspecies, it is important to know the biology, ecology and evolutionary processes of these important pathogens.
AimsThe aim of this study was to propose and validate a low cost identification system for Phytophthora species based on a set of polymorphic microsatellite (SSRs) markers.
MethodsThirty-three isolates representing Phytophthora infestans, Phytophthora andina, Phytophthora sojae, Phytophthora cryptogea, Phytophthora nicotianae, Phytophthora capsici and Phytophthora cinnamomi species were obtained, and 13 SSRs were selected as potentially transferable markers between these species. Amplification conditions, including annealing temperatures, were standardized for several markers.
ResultsA subset of these markers amplified in all species, showing species-specific alleles.
ConclusionsThe adaptability and impact of the identification system in Colombia, an Andean agricultural country where different Phytophthora species co-exist in the same or in several hosts grown together, are discussed.
Phytophthora es el género más importante de los oomicetos, que son patógenos vegetales. Actualmente se han descrito 117 especies de este género, siendo la mayoría invasivas primarias de los tejidos vegetales. Las diferentes especies son agentes causales de enfermedades en una amplia variedad de cultivos y plantas en su medio natural. Con el objetivo de formular estrategias eficientes de control frente a especies de Phytophthora, es primordial conocer la biología, ecología y procesos evolutivos de estos importantes patógenos.
ObjetivosEl objetivo de este estudio fue proponer y validar un sistema de identificación de bajo coste de especies de Phytophthora con una serie de marcadores microsatélites polimórficos (SSR).
MétodosSe obtuvo un total de 33 aislamientos de diferentes especies del género, incluidas Phytophthora infestans, Phytophthora andina, Phytophthora sojae, Phytophthora cryptogea, Phytophthora nicotianae, Phytophthora capsici y Phytophthora cinnamomi. Como marcadores potencialmente transferibles entre estas especies del género Phytophthora se seleccionaron 13 microsatélites, y las condiciones de amplificación, incluidas las temperaturas de alineamiento, se estandarizaron para varios marcadores.
ResultadosUn subgrupo de estos marcadores microsatélites se amplificó en todas las especies mostrando alelos específicos de especie.
ConclusionesSe describen la adaptabilidad e influencia del sistema de identificación en un país agrícola andino como Colombia, donde coexisten diferentes especies de Phytophthora en el mismo huésped o en diversos huéspedes cultivados al mismo tiempo.
Strains belonging to the genus Phytophthora can be found in a high variety of ecological habitats. Among the 117 Phytophthora species described, the most recognized has been Phytophthora infestans, the causal agent of the late blight of solanaceous plant species, which can devastate potato and tomato crops.12 However, Phytophthora infestans is not the only devastating species in this genus. Phytophthora sojae, a pathogen on soybean and some wild flowers, has caused $200 million in annual losses in the United States.38Phytophthora ramorum, the causal agent of oak death, able to infect more than a hundred plant species, has also been extensively studied.20,25 Other Phytophthora species with broad host ranges which have not been widely studied in the Andean region, but that also cause extensive damage on crops and wild plant species comprise Phytophthora capsici that affects cucurbits and causes significant losses on pumpkin crops, and also affects red and green peppers, and ornamental plants2; Phytophthora cinnamomi that affects avocado crops causing root rot22 and has been reported affecting Eucalyptus and wild plants pathogen in Australia and South Africa.30,37Phytophthora nicotianae has been reported in tomato,7 citrus,32 tobacco and wood ornamentals. And finally, Phytophthora cryptogea has been described in many stems in trees, shrubs, ornamental plants and whitlow chicory (Cichorium intybus L.).3
In the Andean region, crops such as potato and tomato are an important agricultural activity.8,9 In addition to potato (Solanum tuberosum and Solanum phureja) and tomato (Solanum lycopersicum), other members of the family Solanaceae are hosts of P. infestans. Over the last decade, several species of Andean exotic fruit have become increasingly important in Colombia for both domestic consumption and the international export market. The most important are Physalis peruviana (cape gooseberry), a herbaceous perennial plant; Solanum betaceum (tree tomato); and Solanum quitoense (lulo or naranjilla), and all of them are hosts of at least one species of Phytophthora.40
Despite the economic impact of diseases caused by species of the genus Phytophthora, studies aiming at understanding the diseases caused by different Phytophthora species have not been reported for the Andean region, with the exception of P. infestans. Species such as P. capsici, P. cinnamomi and P. nicotianae are presumed to be present in the fields but there are no studies addressing this issue. This underscores the importance of developing strategies to evaluate the distribution of these pathogens in several countries.
Several methods that include phenotypic and genotypic markers have been used to identify and to study the population, evolution, genetics, and epidemiology of Phytophthora. Simple sequence repeats (SSR) are well-characterized PCR-based and codominant markers.12 SSR have been used for the identification of mycorrhizal fungi26 and for genetic and population studies of Plasmopara viticola18; P. cinnamomi, P. infestans, P. sojae and P. ramorum.13,24,34 These markers are very helpful for genetic population analysis because they provide a good taxonomic resolution for the analysis of individual isolates in a population, and phylogenetic relationships between related taxonomic groups.12
Microsatellites are DNA simple sequence repeats of one to six base pairs, present in prokaryotic and eukaryotic genomes, and differ from other types of DNA sequences in their high degree of polymorphism derived mainly from variability in length. Besides genetic variation, microsatellite have a high level of heterozygosity, and the presence of multiple alleles.15
One limitation of SSRs is that flanking regions of these sequences have to be known for primer design, and generally primers are designed based on sequences of a certain species. Recently, thanks to the genomics and bioinformatics resources that have been developed, it is useful to evaluate the transferability of previously designed SSRs markers in different species, as the studies performed by Garnica et al.16 and Schena.36 Garnica et al.16 characterized the SSRs from expressed sequences and generated a database (http://bioinf.ibun.unal.edu.co/phytossr/) in which transferable primers between three Phytophthora species were identified.
The aim of this study is to propose and validate a low cost identification system, for seven Phytophthora species based on a set of polymorphic microsatellites markers, and to create a reproducible and reliable identification code for the selected species. These molecular markers will be useful to diagnose and monitor important plant pathogens. Most importantly, the identification system will strengthen the plant health inspection services in Andean countries.
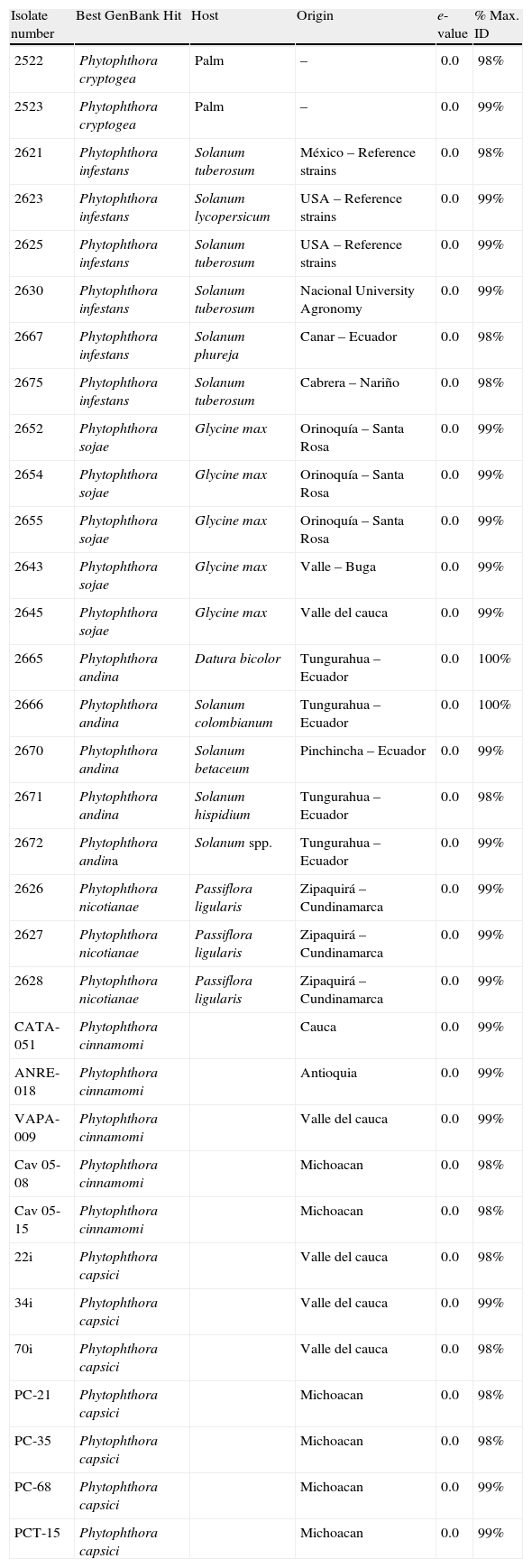
Materials and methodsPhytophthora isolatesTwenty-two isolates including P. infestans, P. sojae, Phytophthora andina, P. cryptogea, and P. nicotianae from different hosts were obtained from the LAMFU culture collection. In addition, three P. capsici isolates were donated by Juan Germán Muñoz from Universidad Nacional-Palmira, three P. cinnamomi isolates were donated by Elizabeth Alvárez from the International Center for Tropical agriculture (CIAT), and finally, three P. capsici, and two P. cinnamomi were donated by Silvia Fernández from IIAF-Michoacán San Nicolás de Hidalgo University (Table 1).
Phytophthora spp. strains collection used in this study. Best GenBank Hit refers to the results obtained from the Blastn search against GenBank nr database that identified each Phytophthora strain. Host refers to the plant where each strain was isolated and origin the place where the isolation was obtained.
| Isolate number | Best GenBank Hit | Host | Origin | e-value | % Max. ID |
| 2522 | Phytophthora cryptogea | Palm | – | 0.0 | 98% |
| 2523 | Phytophthora cryptogea | Palm | – | 0.0 | 99% |
| 2621 | Phytophthora infestans | Solanum tuberosum | México – Reference strains | 0.0 | 98% |
| 2623 | Phytophthora infestans | Solanum lycopersicum | USA – Reference strains | 0.0 | 99% |
| 2625 | Phytophthora infestans | Solanum tuberosum | USA – Reference strains | 0.0 | 99% |
| 2630 | Phytophthora infestans | Solanum tuberosum | Nacional University Agronomy | 0.0 | 99% |
| 2667 | Phytophthora infestans | Solanum phureja | Canar – Ecuador | 0.0 | 98% |
| 2675 | Phytophthora infestans | Solanum tuberosum | Cabrera – Nariño | 0.0 | 98% |
| 2652 | Phytophthora sojae | Glycine max | Orinoquía – Santa Rosa | 0.0 | 99% |
| 2654 | Phytophthora sojae | Glycine max | Orinoquía – Santa Rosa | 0.0 | 99% |
| 2655 | Phytophthora sojae | Glycine max | Orinoquía – Santa Rosa | 0.0 | 99% |
| 2643 | Phytophthora sojae | Glycine max | Valle – Buga | 0.0 | 99% |
| 2645 | Phytophthora sojae | Glycine max | Valle del cauca | 0.0 | 99% |
| 2665 | Phytophthora andina | Datura bicolor | Tungurahua – Ecuador | 0.0 | 100% |
| 2666 | Phytophthora andina | Solanum colombianum | Tungurahua – Ecuador | 0.0 | 100% |
| 2670 | Phytophthora andina | Solanum betaceum | Pinchincha – Ecuador | 0.0 | 99% |
| 2671 | Phytophthora andina | Solanum hispidium | Tungurahua – Ecuador | 0.0 | 98% |
| 2672 | Phytophthora andina | Solanum spp. | Tungurahua – Ecuador | 0.0 | 99% |
| 2626 | Phytophthora nicotianae | Passiflora ligularis | Zipaquirá – Cundinamarca | 0.0 | 99% |
| 2627 | Phytophthora nicotianae | Passiflora ligularis | Zipaquirá – Cundinamarca | 0.0 | 99% |
| 2628 | Phytophthora nicotianae | Passiflora ligularis | Zipaquirá – Cundinamarca | 0.0 | 99% |
| CATA-051 | Phytophthora cinnamomi | Cauca | 0.0 | 99% | |
| ANRE-018 | Phytophthora cinnamomi | Antioquia | 0.0 | 99% | |
| VAPA-009 | Phytophthora cinnamomi | Valle del cauca | 0.0 | 99% | |
| Cav 05-08 | Phytophthora cinnamomi | Michoacan | 0.0 | 98% | |
| Cav 05-15 | Phytophthora cinnamomi | Michoacan | 0.0 | 98% | |
| 22i | Phytophthora capsici | Valle del cauca | 0.0 | 98% | |
| 34i | Phytophthora capsici | Valle del cauca | 0.0 | 99% | |
| 70i | Phytophthora capsici | Valle del cauca | 0.0 | 98% | |
| PC-21 | Phytophthora capsici | Michoacan | 0.0 | 98% | |
| PC-35 | Phytophthora capsici | Michoacan | 0.0 | 98% | |
| PC-68 | Phytophthora capsici | Michoacan | 0.0 | 99% | |
| PCT-15 | Phytophthora capsici | Michoacan | 0.0 | 99% |
Cultures of P. infestans, P. andina, P. capsici, P. cinnamomi, P. cryptogea and P. nicotianae were grown at 19°C for 12–15 days on rye agar, except for P. sojae that was grown on V8 medium. Mycelia were then induced on pea broth using two agar plugs and incubated at 18°C. for two weeks. Mycelia were subsequently recovered with filter paper and placed on Eppendorf tubes and frozen at −80°C. Mycelia were lyophilized for two days. DNA extraction procedures were performed following the guidelines proposed by Goodwin.19
Identification of Phytophthora isolatesTo confirm the identification of each Phytophthora species, amplification and further sequencing of the ITS region were performed. Amplification reactions consisted of 2mM MgCl2, 1× Buffer, 0.2μM dNTPs, 0.2μM of primers ITS5 (forward) and ITS4 (reverse),39 1U of Taq polymerase, and 1μL of DNA (50ng) in a 25μL reaction volume. Amplifications were performed on a BioRad thermal cycler (BioRad, Hercules, CA), with an initial denaturation at 96°C for 3min, followed by 35cycles of 96°C for 1min, 55°C for 1min, 72°C for 2min, and a final extension at 72°C for 10min.
Samples were visualized on 1% agarose gel electrophoresis using Quantity One Software (BioRad, Hercules, CA), and sequenced. Resulting sequences were assembled in CLC Work bench (CLCbio, Aarhus, Denmark) and compared with the BLASTn search tool against the GenBank nr database using default parameters.1
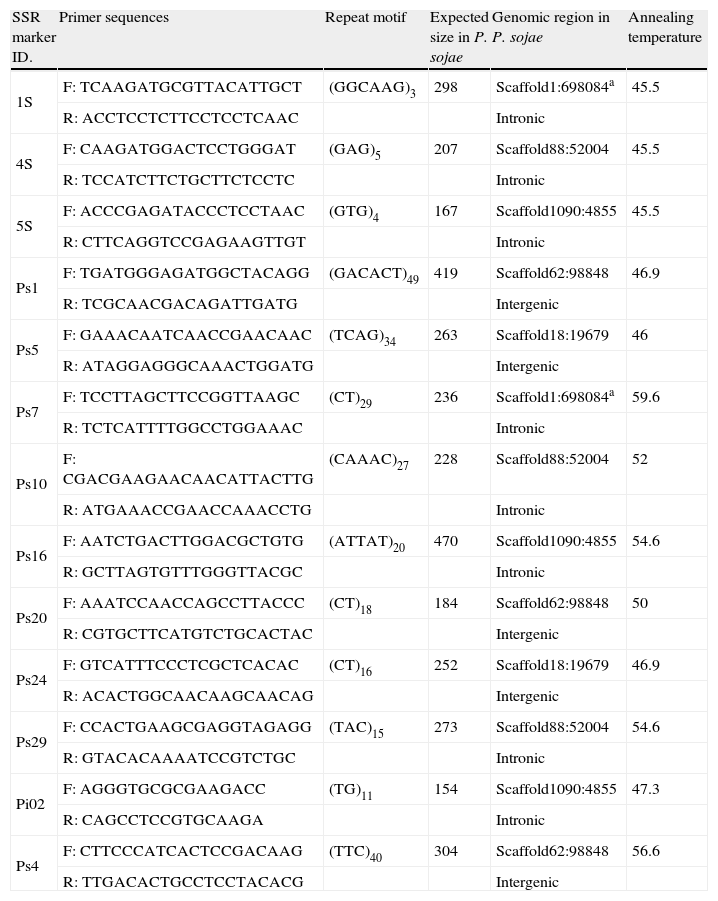
Primer selection and temperature standardizationBased on the SSRs primers designed in previous studies,16,24 13 primer combinations were selected considering the following criteria: (i) specificity for a locus, (ii) transferability among the species assayed by Garnica et al.,16 and (iii) a good quality PCR product.
Gradient PCR, with temperatures between 40 and 60°C was performed with the selected primers (Table 2). Amplification reactions were performed with an initial denaturation at 94°C for 4min, followed by 35cycles of 94°C for 1min, gradient temperature in a range of 40–60°C for 45s, 72°C for 1min, and a final extension at 72°C for 10min. Samples were visualized in 2% agarose gel electrophoresis as described above.
SSRs markers selected for the transferability assay.
| SSR marker ID. | Primer sequences | Repeat motif | Expected size in P. sojae | Genomic region in P. sojae | Annealing temperature |
| 1S | F: TCAAGATGCGTTACATTGCT | (GGCAAG)3 | 298 | Scaffold1:698084a | 45.5 |
| R: ACCTCCTCTTCCTCCTCAAC | Intronic | ||||
| 4S | F: CAAGATGGACTCCTGGGAT | (GAG)5 | 207 | Scaffold88:52004 | 45.5 |
| R: TCCATCTTCTGCTTCTCCTC | Intronic | ||||
| 5S | F: ACCCGAGATACCCTCCTAAC | (GTG)4 | 167 | Scaffold1090:4855 | 45.5 |
| R: CTTCAGGTCCGAGAAGTTGT | Intronic | ||||
| Ps1 | F: TGATGGGAGATGGCTACAGG | (GACACT)49 | 419 | Scaffold62:98848 | 46.9 |
| R: TCGCAACGACAGATTGATG | Intergenic | ||||
| Ps5 | F: GAAACAATCAACCGAACAAC | (TCAG)34 | 263 | Scaffold18:19679 | 46 |
| R: ATAGGAGGGCAAACTGGATG | Intergenic | ||||
| Ps7 | F: TCCTTAGCTTCCGGTTAAGC | (CT)29 | 236 | Scaffold1:698084a | 59.6 |
| R: TCTCATTTTGGCCTGGAAAC | Intronic | ||||
| Ps10 | F: CGACGAAGAACAACATTACTTG | (CAAAC)27 | 228 | Scaffold88:52004 | 52 |
| R: ATGAAACCGAACCAAACCTG | Intronic | ||||
| Ps16 | F: AATCTGACTTGGACGCTGTG | (ATTAT)20 | 470 | Scaffold1090:4855 | 54.6 |
| R: GCTTAGTGTTTGGGTTACGC | Intronic | ||||
| Ps20 | F: AAATCCAACCAGCCTTACCC | (CT)18 | 184 | Scaffold62:98848 | 50 |
| R: CGTGCTTCATGTCTGCACTAC | Intergenic | ||||
| Ps24 | F: GTCATTTCCCTCGCTCACAC | (CT)16 | 252 | Scaffold18:19679 | 46.9 |
| R: ACACTGGCAACAAGCAACAG | Intergenic | ||||
| Ps29 | F: CCACTGAAGCGAGGTAGAGG | (TAC)15 | 273 | Scaffold88:52004 | 54.6 |
| R: GTACACAAAATCCGTCTGC | Intronic | ||||
| Pi02 | F: AGGGTGCGCGAAGACC | (TG)11 | 154 | Scaffold1090:4855 | 47.3 |
| R: CAGCCTCCGTGCAAGA | Intronic | ||||
| Ps4 | F: CTTCCCATCACTCCGACAAG | (TTC)40 | 304 | Scaffold62:98848 | 56.6 |
| R: TTGACACTGCCTCCTACACG | Intergenic |
We also updated the Phytophthora SSR databases16 at http://bioinf.ibun.unal.edu.co/phytossr/including SSR sequences and primers from genomic sequences.
Transferability assayWith the selected temperatures, amplifications were performed using a touchdown PCR, starting the annealing phase at 60°C, until reaching the optimal temperature for each set of primers selected for the SSRs regions. Products were visualized and quantified on 3% high resolution agarose electrophoreses using Quantity One Software (BioRad, Hercules, CA). Gels were run for 3h at 80V using high resolution agarose gels.
Reproducibility of PCR was tested for all the markers with all the isolates in the study. PCR products were run three times in different high resolution agarose gels, and also PCR was performed at least twice with each primer. Also some products were run in acrylamide gels, and visualized by silver staining.27
The amplification products obtained that showed polymorphism within a species were sequenced. Resulting sequences were assembled in CLC Work bench (CLCbio, Aarhus, Denmark) and multiple alignments of the SSRs region were performed.
Data analysisAmplification products obtained with the 13 pair of SSR primers were scored as either present (1) or absent (0). Biodiversity Pro31 was used to calculate a similarity matrix and to construct a dendrogram by Bray–Curtis cluster analysis. Bray–Curtis calculates standard correlation coefficients between all the traits analyzed (in this case the 13 SSRS markers) among the isolates evaluated.5 The dendrogram obtained shows the relationship between the species according to the markers tested, but it did not show phylogenetic relationships.
ResultsPhytophthora species identificationAll the strains were taxonomically confirmed by morphological description, measuring sporangial size (data not shown) and also confirming the ellipsoid to ovoid form of the sporangia for all the species studied, except for P. cryptogea, which was mainly ovoid (Table 1). Sequencing of the ITS region was also performed. The PCR product corresponded to the 900pb expected size,28 and BLASTn analysis of the sequences resulted in hits with e-values of 0.0 and a maximum identity ranging from 98 to 100% corresponding to the ITS region of the different Phytophthora species (Table 1).
Identification system of Phytophthora sp. based on SSRsIn a previous study16 a database containing all the in silico analyses of SSRs derived from EST sequences from P. sojae, P. infestans and P. ramorum, was constructed (http://bioinf.ibun.unal.edu.co/phytossr/). In this database all primers designed for SSRs of the three species are reported, and the online service allows for transferability assays of these markers between the three species. In this study, we uploaded SSRs and primers designed for these regions obtained from entire genomic sequences of P. infestans taking the latest Phytophthora sequence drafts available.
According to the parameters established by Garnica et al.16 for primer selection, primer sets 1S, 4S and 5S designed for SSRs located in intronic regions, primer set for locus Pi02 by Lees et al.24 derived from genomic sequences of P. infestans and primers for PS1, PS4, PS5, PS7, PS10, PS16, PS24, PS29 SSRs located in intergenic regions of P. sojae, were chosen (Table 2).
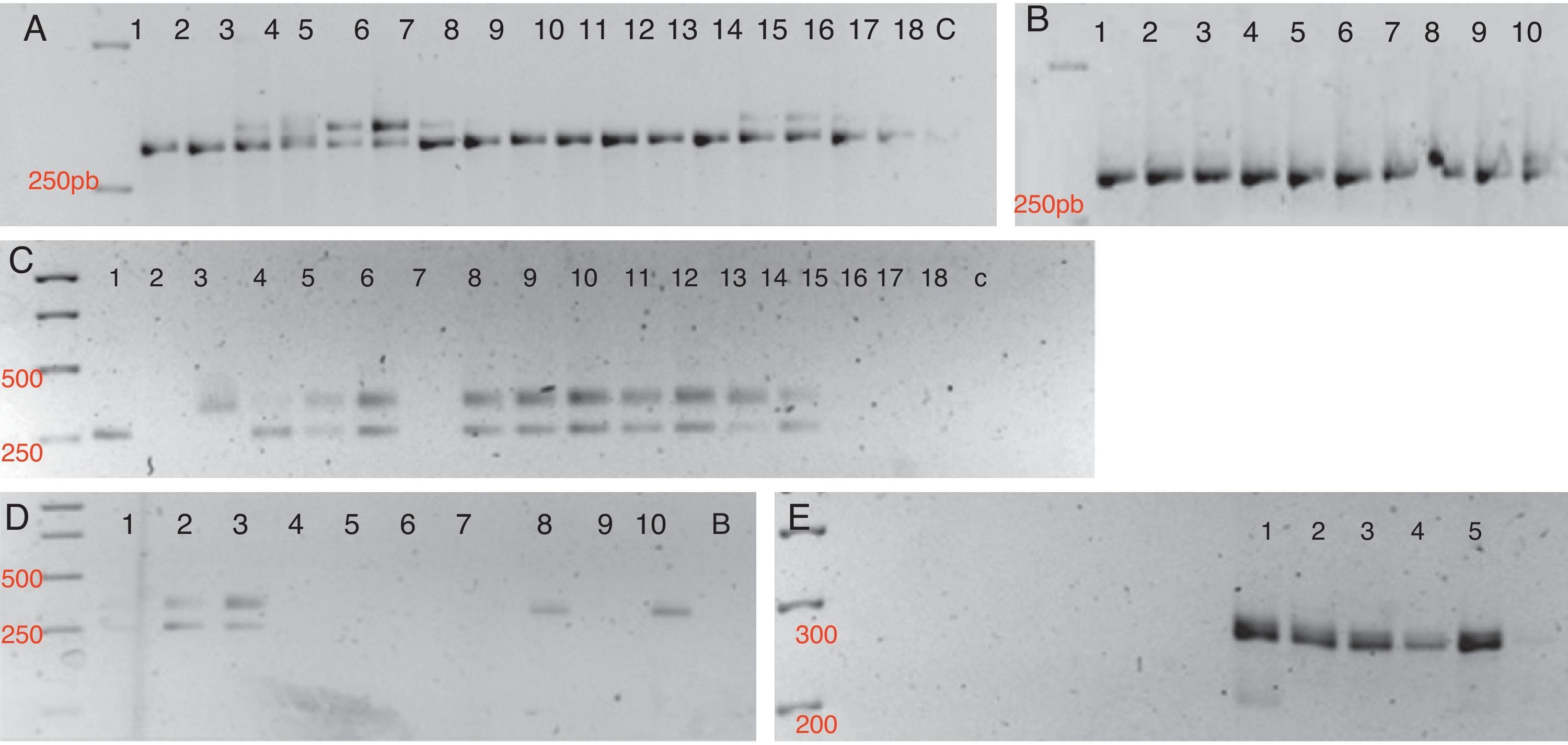
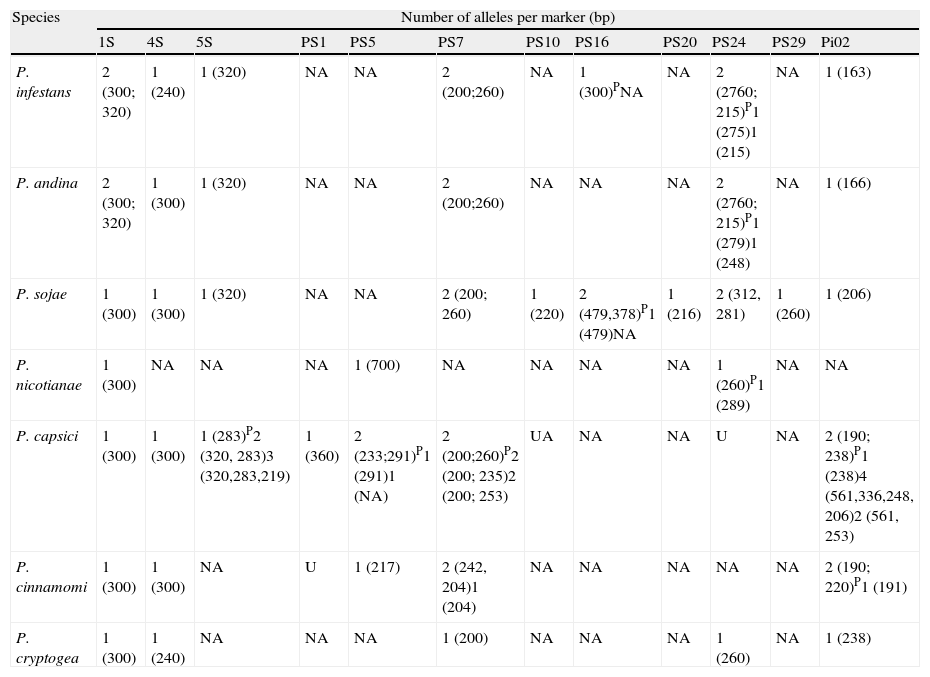
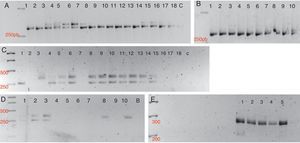
A specific band pattern for each species analyzed was obtained for the thirteen markers tested (Fig. 1). It is also important to note that with this set of loci we could discriminate between the seven Phytophthora species analyzed (Table 3), and that a high polymorphism level is shown within the genus.
Detection of polymorphisms in a 3% high resolution agarose gel. A. 1S SSR Marker. Lanes 1, 2. P. cryptogea; Lanes 3–8. P. infestans; Lanes 9–13. P. sojae; Lanes 14–18. P. andina; C. control. B. 1S SSR Marker. Lanes 1–5. P. capsici; Lanes 5–10. P. cinnamomi.C. Ps7 SSR marker. Lanes 1, 2. P. cryptogea; Lanes 3–7. P. infestans; Lanes 8–12. P. sojae; Lanes 13–15. P. andina; Lanes 16–18. P. nicotianae; C. control. D. Ps7 SSR Marker. Lanes 1–5. P. capsici; Lanes 6–10. P. cinnamomi. E. Ps29 SSR marker. Lanes 1–5. P. sojae.
Number and sizes of alleles per SSRs marker in seven Phytophthora species.
| Species | Number of alleles per marker (bp) | |||||||||||
| 1S | 4S | 5S | PS1 | PS5 | PS7 | PS10 | PS16 | PS20 | PS24 | PS29 | Pi02 | |
| P. infestans | 2 (300; 320) | 1 (240) | 1 (320) | NA | NA | 2 (200;260) | NA | 1 (300)PNA | NA | 2 (2760; 215)P1 (275)1 (215) | NA | 1 (163) |
| P. andina | 2 (300; 320) | 1 (300) | 1 (320) | NA | NA | 2 (200;260) | NA | NA | NA | 2 (2760; 215)P1 (279)1 (248) | NA | 1 (166) |
| P. sojae | 1 (300) | 1 (300) | 1 (320) | NA | NA | 2 (200; 260) | 1 (220) | 2 (479,378)P1 (479)NA | 1 (216) | 2 (312, 281) | 1 (260) | 1 (206) |
| P. nicotianae | 1 (300) | NA | NA | NA | 1 (700) | NA | NA | NA | NA | 1 (260)P1 (289) | NA | NA |
| P. capsici | 1 (300) | 1 (300) | 1 (283)P2 (320, 283)3 (320,283,219) | 1 (360) | 2 (233;291)P1 (291)1 (NA) | 2 (200;260)P2 (200; 235)2 (200; 253) | UA | NA | NA | U | NA | 2 (190; 238)P1 (238)4 (561,336,248, 206)2 (561, 253) |
| P. cinnamomi | 1 (300) | 1 (300) | NA | U | 1 (217) | 2 (242, 204)1 (204) | NA | NA | NA | NA | NA | 2 (190; 220)P1 (191) |
| P. cryptogea | 1 (300) | 1 (240) | NA | NA | NA | 1 (200) | NA | NA | NA | 1 (260) | NA | 1 (238) |
NA: no amplification; P: polymorphisms found at intraspecific level; U: unspecified amplification.
At the intraspecific level, we found low levels of polymorphism in 1S, 4S, 5S, PS7 and Pi 02 loci, for P. sojae, P. infestans, P. andina and P. nicotianae, obtaining a very similar banding pattern for these species. Nevertheless, with locus PS24 we found intraspecific polymorphisms for P. infestans and P. andina. Phytophthora capsici and P. cinnamomi strains showed high levels of polymorphism with markers 5S, PS7 and Pi02.
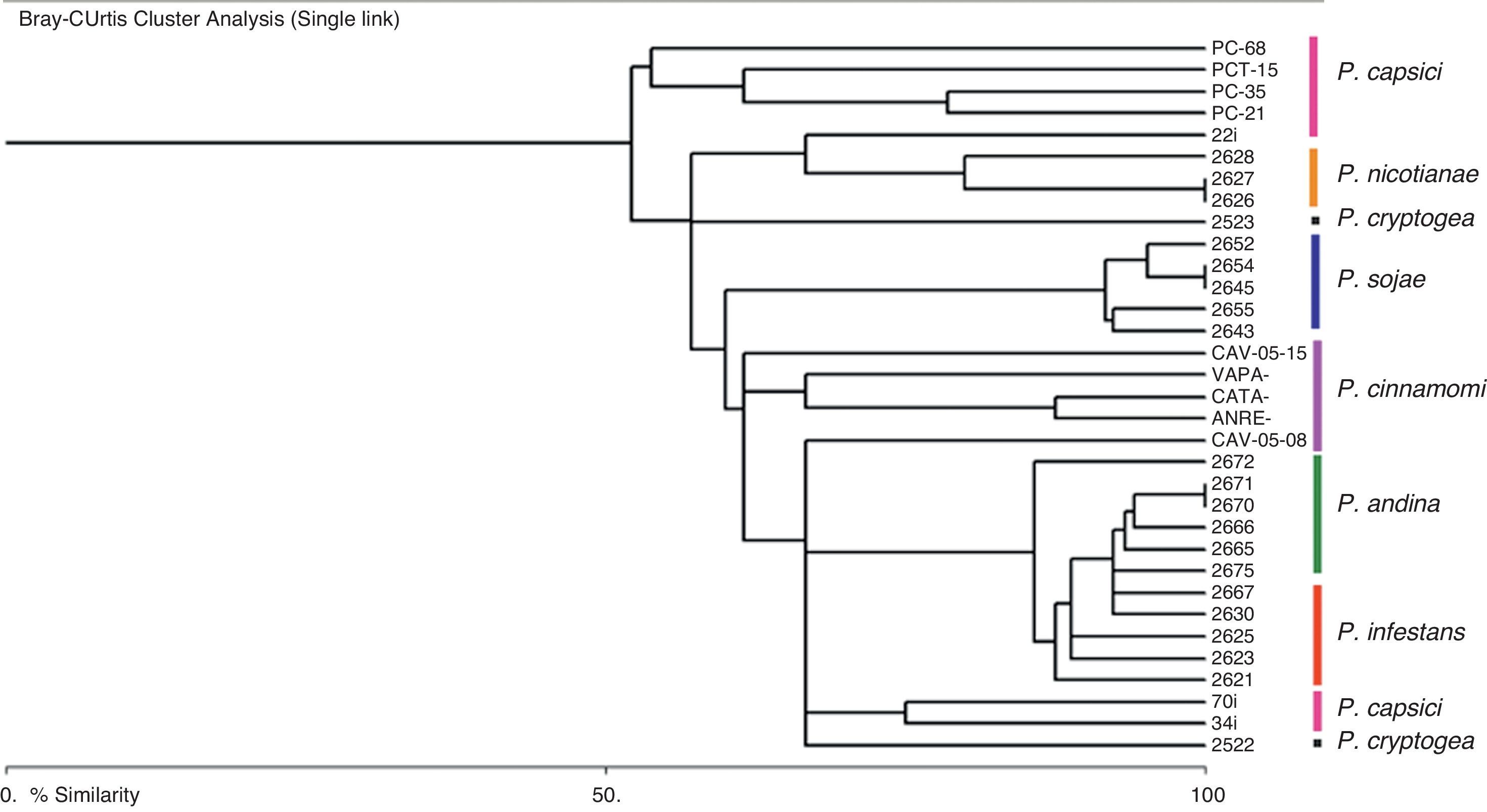
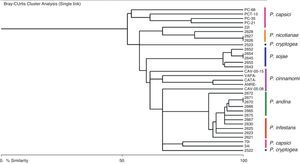
The dendrogram (Fig. 2) derived of the Bray–Curtis cluster analysis showed that the strains belonging to P. infestans, P. andina, P. sojae, P. nicotianae and P. cinnamomi were sorted in the same cluster respectively, and the strains of P. capsici and P. cryptogea were not. It is also interesting that in the case of P. capsici, the strains from Mexico are clustered together, whereas two strains from Colombia form a different cluster, suggesting a geographically dependent intraspecific variation, as stated by Bowers et al.6 Also, in this dendrogram we confirmed that P. infestans, and P. andina, are closely related, and we could differentiate between these species with the loci Ps16, and 4S (Table 3). To differentiate P. sojae from all the others species, loci PS20, and PS29 can be used, as they only amplify in this species. For P. nicotianae loci 1S, PS5 and PS24, for P. capsici locus PS1, for P. cinnamomi locus PS5 and for P. cryptogea the set of loci 1S, 4S, PS7, PS24, and Pi02 could be used, although more strains from P. cryptogea will be needed to confirm species delimitation. In terms of the band sizes obtained with these amplifications, all of them were higher than the expected sizes obtained in our previous study.16
In order to prove that the amplification products obtained had indeed the SSRs sequence, some products were sequenced. We found that amplification products of 1S and 4S markers contained the SSRs motif. Only for strains of P. sojae and P. capsici two repetitions of the 4S SSR were found. On the other hand, the amplification products of Pi02 contained the SSR (TG)11, but with three and two more repetitions of the motif for P. infestans and P. andina, respectively, when compared with the sequence of P. sojae. For the PS24 marker, the SSR (CT)16 was only found in P. sojae strains.
Many population studies that use neutral molecular markers, such as SSRs, RFLPs and SNPs have been developed using acrylamide gels, because the power of resolution is considered higher and more accurate than those of agarose gels. To prove that the SSRs analyses visualized on high resolution agarose gel had a good resolution power and that the alleles obtained were not overlapped, we visualized the products of PS24 and 1S markers on acrylamide gels. The band patterns were similar for both kinds of gels, and the alleles observed in high-resolution agarose were not overlapping.
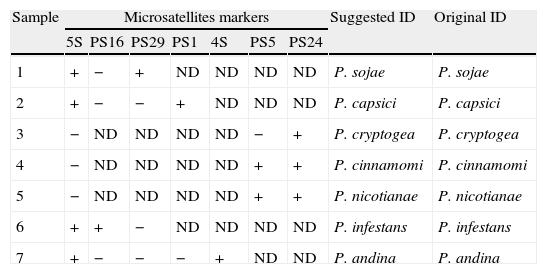
In addition, to prove that the method proposed is reliable, a blind test was performed. Seven isolates that were previously identified morphologically (Table 4) were analyzed with the SSRs set. First, we tested the primer pair for the 5S locus, which allowed differentiating species that had an amplification product such as P. andina, P. infestans, P. capsici and P. sojae from P. nicotianae, P. cinnamomi and P. cryptogea, which do not amplified. From the seven target species to identify, samples 1, 2, 6 and 7 had an amplification product, whereas samples 3, 4 and 5 did not have it. With this result, samples 1, 2, 6 and 7 were tested with primer PS16, and an amplification product was obtained for sample 6 only, the rest of these samples were tested with primer PS29, obtaining a band only for sample 1. The samples 2 and 7 were tested with primer PS1, where an amplification product was obtained for sample 2, and finally sample 7 was tested with primer 4S, obtaining an amplification product of 290 base pairs. With this result it can be suggested that sample 1 is P. sojae, 2 is P. capsici, 6 is P. infestans and 7 is a P. andina. Samples 3, 4 and 5 were tested with primer PS5, whereas amplification was obtained only for samples 4 and 5. Finally marker PS24 was tested and amplification products were obtained for the three samples tested. With these results, sample 3 would be classified as P. cryptogea, sample 4 as P. cinnamomi and sample 5 as P. nicotianae. An independent analyst performed the test confirming the obtained results.
Blind test performed to confirm the usefulness of the SSRs set to identify Phytophthora species.
| Sample | Microsatellites markers | Suggested ID | Original ID | ||||||
| 5S | PS16 | PS29 | PS1 | 4S | PS5 | PS24 | |||
| 1 | + | − | + | ND | ND | ND | ND | P. sojae | P. sojae |
| 2 | + | − | − | + | ND | ND | ND | P. capsici | P. capsici |
| 3 | − | ND | ND | ND | ND | − | + | P. cryptogea | P. cryptogea |
| 4 | − | ND | ND | ND | ND | + | + | P. cinnamomi | P. cinnamomi |
| 5 | − | ND | ND | ND | ND | + | + | P. nicotianae | P. nicotianae |
| 6 | + | + | − | ND | ND | ND | ND | P. infestans | P. infestans |
| 7 | + | − | − | − | + | ND | ND | P. andina | P. andina |
ND: not determined.
Microsatellites are highly useful for genotypic identification and population studies because of their high throughput, robust and flexible nature, as well as their suitability for rigorous genetic analysis. They have also been broadly applied to diverse analyses and can be classified as safe.12 In this study we propose a new identification system for seven Phytophthora species with transferable SSRs markers. All the species analyzed could be differentiated using the set of SSRs primers selected. Our results also showed that the SSRs could be an ideal marker system for the genetic analysis of the Phytophthora species analyzed, suggesting that these markers can be employed for the entire genus. Previous studies in oomycetes reported the use of microsatellites at the intraspecific level in P. cinnamomi,13P. ramorum,34P. infestans24 and Plasmopara viticola,17 but the use of these molecular markers at interspecific level has just recently been exploited36 and could be a very valuable tool for inspection services in Andean countries, where a diversity of hosts for these species is cultivated.
Since 2008, tools such as the Phytophthora database (http://www.phytophthoradb.org/) have contributed to share data among the Phytophthora research community. This database contains reliable sequences of the genus, including ribosomal DNA (ITS region and LSU), several nuclear genes, and the mitochondrial encoded coxII and spacer region between coxI and coxII33 of most of the species described to date, as well as tools for bioinformatics analyses. It also has contributed with the significant increase of Phytophthora species, from 90 species described by 2008,21 to up to 117 described species to date.29
Currently diagnostics and identification of species are based on sequencing, and for the Phytophthora genus, sequence-based identification tools have been recently proposed by Ristaino35 and Grünwald et al.21 The site http://phytophthora-id.org/contains curated ITS and cox spacer sequences from an extensive collection of Phytophthora spp.21 Although the ITS region has been used for species identification,10,11,14,23 some closely related Phytophthora species that are prevalent in the Andean region can not be differentiated, such as P. infestans and P. andina. Additionally, in Colombia the sequencing process is still expensive and thus a limiting factor for the sequence-based identification.
It is important to notice that all the experimental procedures carried in this study were initially based on the in silico data generated by Garnica et al.,16 and that the use of genomic sequences, EST and all the information available in different databases for Phytophthora spp. such as http://www.broad.mit.edu/ for P. infestans, and http://genome.jgi-psf.org/ for P. sojae, P. ramorum, and P. capsici, can be applied to develop population studies that are helpful to understand the evolution, diversity, population structure and epidemiology of these plant pathogens. We also contributed to all this in silico information for the Phytophthora genus by creating and now updating an SSR database with information about transferability (http://bioinf.ibun.unal.edu.co/phytossr/).
To develop this kind of identification approach in the national plant health services, the economic factor is a strong determinant in the selection of the best methodology to achieve an accurate analysis at a low cost. We are therefore proposing an identification system of Phytophthora species that comprises all these characteristics: low cost, accuracy and applicability in developing countries. In the case of Colombia, for example, the costs of all the reagents needed for an acrylamide gel are up to US$ 11.96 for a gel with 40 samples, whereas for a high-resolution agarose the costs are up to US$ 2.66 for a gel with 20 samples.
With the set of primers employed we could obtain amplification products for all the species tested, and allele patterns that allowed discrimination between species, even closely related ones. However, some of the amplified products did not contain the SSRs in any of the Phytophthora species analyzed, such as the marker 1S. This result was also observed by Garnica et al.16 with Phytophthora sojae. Although we did not obtain the SSRs in the sequence, the primers were transferable for all Phytophthora species tested. This can be a useful marker for species delimitation in the genus, given that the amplification product showed polymorphism in the seven species tested.
Primers for PS7 and PS24 loci, derived from intergenic regions of P. sojae, amplified in all the tested species, suggesting that the flanking regions of the SSR could be conserved in the genus. Nonetheless, the SSR sequence corresponding to PS24 was only found in P. sojae, probably because the PS24 and PS7 SSRs are dinucleotide repeats, and according to Schena36 these were the less frequent SSRs for P. infestans, P. ramorum and P. sojae. On the other hand, for P. capsici and P. cinnamomi nonspecific amplifications were observed. This can be due to the fact that multiple copies of the flanking region can be present in the genomes.
The primer pairs for Pi02 were designed from a genomic region of P. infestans and showed transferability in all the species, but the SSRs sequenced were present only in P. infestans and P. andina and different number of repeats was observed, showing that different alleles are present for this locus across the genus. Lees et al.24 also showed that Pi02, and nine SSRs markers assayed in P. infestans had a variable number of SSR repeats for each marker, being ideal markers for intraspecific identification. Although, more P. infestans and P. andina isolates should be tested for the Pi02 locus, this region could be useful for the molecular differentiation of these species which is an epidemiological issue in the southwestern region of our country, due to the hybrid origin of P. andina.8,20 Primers PS20 and PS29 from intergenic regions were specific for P. sojae, showing that some regions are highly conserved within the species and that for some markers transferability is difficult to achieve particularly when the genetic distance increases inside the genus.
Our results based on SSRs markers agree with the latest phylogeny of the genus.4 We showed in the SSR-derived dendrogram that species such as P. andina and P. infestans, are closely related as well as P. sojae and P. cinnamomi. In the latest phylogeny, P. andina and P. infestans belong to clade 1C, and P. sojae and P. cinnamomi to clade 7.4 Also, this dendrogram showed that a high diversity is presumed within the genus. Although an amplification pattern was obtained for most of the species, the SSRs are not necessarily present in all the species. Nevertheless, it is important to mention that relationships among the species in the dendrogram are useful for species identification and basic genetic trends but not for phylogenetic studies.
Molecular studies that include nuclear, ribosomal DNA and multiloci information have been employed to resolve relationships in the genus Phytophthora.4 The ITS region has been employed in several studies10,11,23 but this region does not provide enough reliability and discrimination power to differentiate between close species such as the ones belonging to clade 1c. The introduction of a novel identification system such as SSRs that allows the diagnostic and detection of Phytophthora species is very useful because it enhances sample throughput, yet it can be used in developing countries that do not have enough resources for identification systems such as sequencing, and it will contribute to monitor the epidemiology of these devastating pathogens that have a world wide distribution.
Conflict of interestThe authors declare they have no conflict of interest.