The extent to which commercially available nucleic acid extraction platforms impact the magnitude of Cytomegalovirus (CMV) DNA loads measured in plasma specimens by 1st WHO standard-normalized real-time PCR assays is uncertain.
MethodsThis retrospective study compares the performance of Abbott m2000sp, Qiagen QIAsymphony SP, and KingFisher Flex platforms using plasma samples from allogeneic hematopoietic stem cell transplant recipients and plasma spiked with the CMV AD169 strain. The Abbott RealTime CMV PCR assay was used for CMV DNA quantitation.
ResultsMaximum differences in CMV DNA loads quantified in plasma from 11 allo-HSCT and spiked plasma over a wide range of viral DNA concentrations (2.0–4.0 log10 IU/ml) were ≤0.5 log10 IU/ml.
ConclusionsThe CMV DNA extraction efficiency of the platforms evaluated varies. The impact of these variations on CMV DNA loads quantified in plasma may not be clinically relevant.
Se desconoce si el uso de distintas plataformas de extracción de ácidos nucleicos afecta la magnitud de las cargas de ADN de citomegalovirus (CMV) cuantificadas mediante PCR en tiempo real normalizadas al primer estándar de la OMS.
MétodosComparamos retrospectivamente las plataformas Abbott m2000sp, Qiagen QIAsymphonySP y KingFisher Flex utilizando muestras de plasma de receptores de trasplante alogénico hematopoyético (alo-TPH) y plasma inoculado con la cepa CMV AD169. Las cargas virales se cuantificaron mediante el ensayo Abbott RealTime CMV PCR.
ResultadosLas diferencias máximas en las cargas cuantificadas en plasma de 10 alo-TPH y plasma inoculado, en un rango amplio de concentraciones (2,0 a 4,0 log10 UI/ml) fueron ≤0,5 log10 UI/ml.
ConclusionesLa eficiencia de extracción de ADN de CMV de las plataformas analizadas varía; sin embargo, el impacto de estas variaciones en las cargas de ADN del CMV cuantificadas en plasma podría no ser clínicamente relevante.
Monitoring Cytomegalovirus (CMV) DNA load in blood (plasma or whole blood) is a cornerstone in the management of CMV infection in transplant recipients.1 Normalization of CMV DNA loads, quantified by different commercially available nucleic acid amplification testing (NAAT) platforms, to the 1st WHO international standard2 has led to improved agreement in CMV DNA load values across assays; yet, variability persists.3 The size of the PCR amplicon generated largely explains such variability4,5 as CMV DNA present in blood, most notably in plasma, is highly fragmented6–8; nevertheless, other factors may also contribute. In this sense, the efficiency of nucleic acid extraction platforms was shown to have a sizeable impact on the magnitude of CMV DNA loads measured by different real-time PCRs prior to their normalization to the 1st WHO international standard.9,10 Moreover, by using small-sized synthetic oligonucleotides and a droplet digital PCR assay for CMV DNA quantitation, Cook et al.11 showed that nucleic acid yields vary widely across extraction platforms, particularly for small DNA fragments (≤100bp). Refined magnetic separation-based rapid nucleic acid extraction technology has been increasingly incorporated, both in automated nucleic acid extraction systems and high-throughput platforms, combining nucleic acid extraction and NAAT.12 There is scarce information13 regarding how the performance of recent-generation nucleic acid extraction platforms impacts the magnitude of CMV DNA loads measured in plasma specimens by 1st WHO standard-normalized real-time PCR assays. Here, we addressed this issue by measuring CMV DNA loads in plasma from allogeneic hematopoietic stem cell transplant recipients (allo-HCT) after nucleic acid extraction in three widely used magnetic separation-based nucleic acid extraction platforms currently in use in our laboratory.
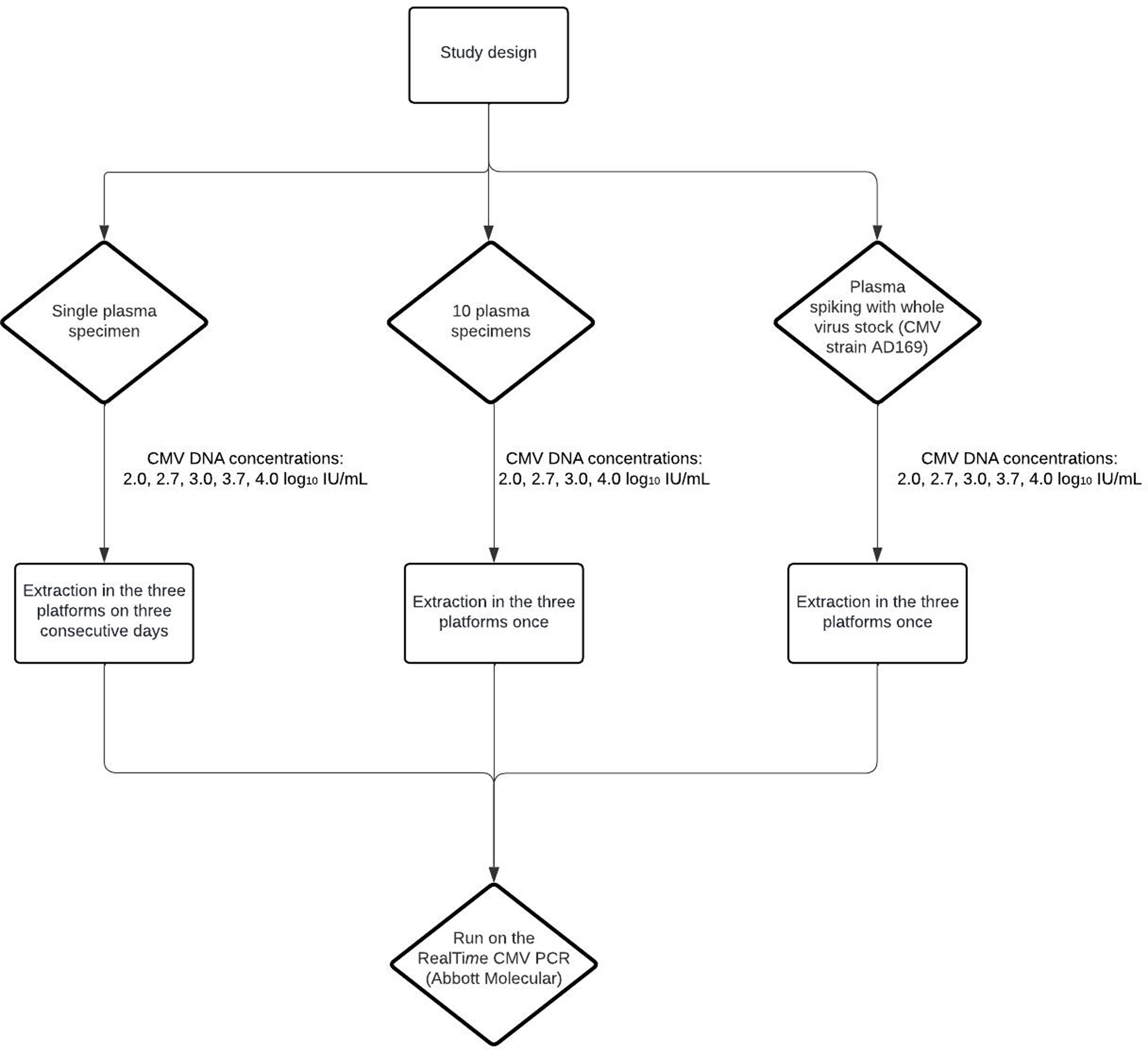
Material and methodsA total of 11 cryopreserved (−80°C for less than one year) plasma specimens from 11 allo-HCT patients who developed CMV DNAemia were retrieved for the analyses detailed below. CMV DNA load in these specimens had been routinely quantified by the Alinity m CMV assay (Abbott Molecular Inc., Des Plaines, IL, USA). For some experiments, plasma specimens testing negative for CMV DNA by the Alinity m CMV assay were spiked with different concentrations of the CMV AD169 strain. The following nucleic acid extraction platforms were used according to the different manufacturer's instructions: (i) Abbott m2000sp mSample Preparation System DNA; (ii) Qiagen QIAsymphony SP DSP Virus/Pathogen Midi Kit (Hilden, Germany); (iii) KingFisher Flex MagMAX™ Pathogen RNA/DNA Kit (Waltham, Massachusetts, USA). Relevant characteristics of these extraction platforms are shown in Supplementary Table 1. The RealTime CMV PCR (Abbott Molecular), a dual target (UL34 and UL80.5) PCR, with a limit of detection and quantification of 31.4IU/ml (95% CI) was employed for CMV DNA quantitation. CMV DNA loads used for the analyses described below were normalized to the input and elution volumes of each extraction platform as follows: final (corrected) CMV DNA load in IU/ml: measured CMV DNA load in IU/ml×correction factor which is derived from: elution volume/PCR mix volume×units volume/input volume. This only applies to the KingFisher Flex platform. Means and coefficients of variation (CV%) of log10 transformed values, calculated using Excel 2021 (version 18.0) software, are reported throughout the study. The current study was approved by the Ethics Committee of the Hospital Clínico Universitario-INCLIVA (November 2022). The requirement for informed consent was waived by the Ethics Committee.
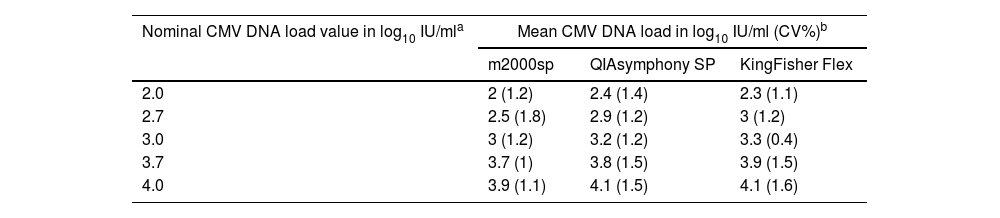
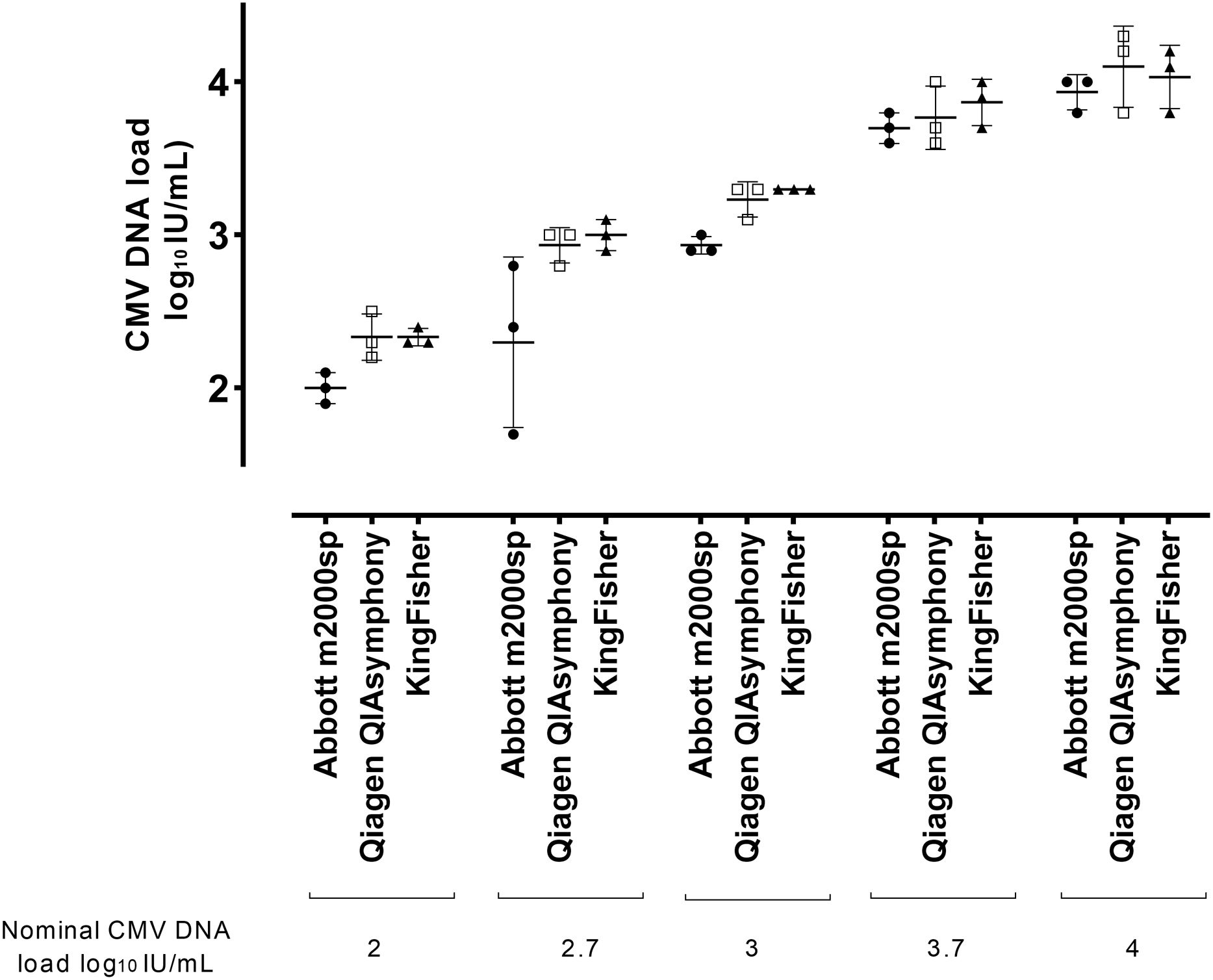
ResultsA scheme of the study design is shown in Supplementary Figure 1. The study initial experiments were performed using a single plasma specimen with a high CMV DNA load, as measured by the Alinity m CMV assay (83,718IU/ml), which was conveniently diluted in pooled CMV DNA-negative plasma specimens, yielding the following CMV DNA concentrations: 2.0, 2.7, 3.0, 3.7 and 4.0 log10 IU/ml. Nucleic acid extraction was carried out in the three platforms on three consecutive days; nucleic acid eluates were then run in singlets on the m2000rt platform (a total of 45 tests). As shown in Table 1, both the QIAsymphony SP and KingFisher Flex, which performed similarly across all CMV DNA concentrations tested (maximum difference, 0.1 log10 IU/ml), were slightly more efficient than the m2000sp platform, according to the magnitude of CMV DNA loads measured. The maximum difference between QIAsymphony SP and m2000sp was 0.40 log10 (at CMV DNA concentrations ≤2.7 log10 IU/ml), and between KingFisher Flex and m2000sp, it was 0.50 log10 (at CMV DNA concentrations of 2.7 log10 IU/ml).
Cytomegalovirus DNA loads quantified by the RealTime CMV PCR assay on a single plasma specimen following nucleic acid extraction using different platforms carried out in three consecutive days.
| Nominal CMV DNA load value in log10 IU/mla | Mean CMV DNA load in log10 IU/ml (CV%)b | ||
|---|---|---|---|
| m2000sp | QIAsymphony SP | KingFisher Flex | |
| 2.0 | 2 (1.2) | 2.4 (1.4) | 2.3 (1.1) |
| 2.7 | 2.5 (1.8) | 2.9 (1.2) | 3 (1.2) |
| 3.0 | 3 (1.2) | 3.2 (1.2) | 3.3 (0.4) |
| 3.7 | 3.7 (1) | 3.8 (1.5) | 3.9 (1.5) |
| 4.0 | 3.9 (1.1) | 4.1 (1.5) | 4.1 (1.6) |
CMV: Cytomegalovirus; CV: coefficient of variation; IU: International Units.
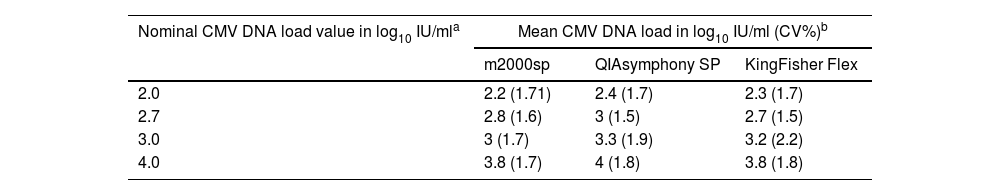
To further assess the impact of the nucleic acid extraction yield of each platform on CMV DNA load values, 10 plasma specimens from 10 allo-HCT recipients with CMV DNA loads ranging from 13,492 to 195,872IU/ml, as quantified by the Alinity m CMV assay, were serially diluted in pooled CMV DNA-negative plasma to achieve nominal viral DNA concentrations of 2.0, 2.7, 3.0, and 4.0IU/ml. Plasma specimens were processed in the three platforms (once) and run on the m2000rt PCR system (in singlets) (a total of 120 tests). As shown in Table 2, and Supplementary Figure 2, the QIAsymphony SP platform was slightly more efficient than the other two systems across all CMV DNA concentrations tested, although maximum differences in CMV DNA loads were ≤0.25 log10 IU/ml. Finally, we spiked pooled CMV DNA-negative plasma with increasing concentrations (2.0, 2.7, 3.0, 3.7 and 4.0IU/ml) of a whole virus stock (CMV strain AD169), whose CMV DNA content had been quantified by the Alinity m CMV assay. Plasma specimens were processed in the three platforms on three consecutive days and then run in singlets on the m2000rt system (n=36 tests). As shown in Supplementary Table 2, overall, maximum differences across platforms did not exceed 0.5 log10 IU/ml, with the QIAsymphony SP platform performing discretely better at some but not all CMV DNA concentrations.
Cytomegalovirus DNA loads quantified by the RealTime CMV PCR assay in plasma from 10 allogeneic hematopoietic stem cell transplant recipients following nucleic acid extraction using different platforms.
| Nominal CMV DNA load value in log10 IU/mla | Mean CMV DNA load in log10 IU/ml (CV%)b | ||
|---|---|---|---|
| m2000sp | QIAsymphony SP | KingFisher Flex | |
| 2.0 | 2.2 (1.71) | 2.4 (1.7) | 2.3 (1.7) |
| 2.7 | 2.8 (1.6) | 3 (1.5) | 2.7 (1.5) |
| 3.0 | 3 (1.7) | 3.3 (1.9) | 3.2 (2.2) |
| 4.0 | 3.8 (1.7) | 4 (1.8) | 3.8 (1.8) |
CMV: Cytomegalovirus; CV: coefficient of variation; IU: International Units.
CMV DNA in plasma from transplant recipients is mostly unprotected and highly fragmented,6–8 with fragments ≤100bp representing the largest fraction of viral DNA content.8 The efficiency of different commercially available extraction platforms to extract CMV DNA fragments of 50 or 100bp was shown to vary widely across systems11; nevertheless, in this study, specimens spiked with synthetic oligonucleotides analyzed via droplet digital PCR (ddPCR) for CMV DNA quantitation were used in the experiments. ddPCR is based on the isolated amplification of thousands of individual DNA molecules simultaneously, with each molecule compartmentalized in a droplet. The presence of amplified product in each droplet is indicated by a fluorescent signal, and the proportion of positive droplets allows the precise quantification of a given sequence in the absence of quantitation standards.14 Here, we examined whether three magnetic separation-based automated nucleic acid extraction platforms used for clinical plasma specimens significantly impacted CMV DNA loads, as measured by a 1st WHO standard-normalized real-time PCR assay. Two major observations were made. First, there were minimal differences, although sizeable, in the performance of the three extraction platforms when clinical plasma specimens from allo-HCT recipients were tested; the QIAsymphony SP platform was slightly more efficient. Differences in CMV DNA loads measured following extraction with the three platforms were overall ≤0.5 log10 IU/ml, and were more pronounced at low CMV DNA concentrations (≤2.7 log10 IU/ml). This observation should be taken into consideration when plasma specimens need to be pre-diluted (i.e. in neonates) to achieve the minimum volume required for extraction according to the respective manufacturer. This figure aligns with previously published data. In effect, Kim et al.13 found a mean difference of −0.32 log10 copies/ml between the QIAsymphony RGQ and QIAcube systems using the Artus CMV QS RGQ and RG assays for CMV DNA quantification. In turn, Bravo et al.9 reported CMV DNA loads quantified by the RealTime CMV PCR assay to differ by ≤0.4 log10 copies/ml following extraction with the m2000sp system, the High Pure viral nucleic acid kit on the COBAS AmpliPrep system (Roche Diagnostics, Mannheim, Germany), and the EZ1 Virus 2.0 kit (Qiagen, Valencia, CA) on the BioRobot EZ1. In this context, a difference of <0.5 log10 IU/ml across CMV NAAT assays is not considered clinically relevant, although this extent is debatable. Second, all extraction platforms evaluated appeared to perform comparably with clinical plasma specimens (highly fragmented CMV DNA) and plasma spiked with whole virus (CMV DNA mostly protected and thus non-fragmented). The main limitation of the current study is the scarce number of clinical specimens tested. In summary, our data indicated that, although the CMV DNA extraction efficiency of commercially available platforms varies, the potential impact of these variations on CMV DNA loads quantified in clinical plasma specimens by 1st WHO standard-normalized real-time PCR assays may be clinically irrelevant. Naturally, our assumption cannot be extended to extraction platforms and real-time PCR assays other than those evaluated herein.
FundingThis research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Authors’ contributionsÁngela Sánchez-Simarro, Eliseo Albert, Paula Michelena, and Estela Giménez: Methodology, investigation, formal analysis, data curation, and writing review & editing. David Navarro: Conceptualization, investigation, formal analysis, and writing the original draft.
Competing interestsThe authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Ángela Sánchez-Simarro (PFIS Contract; FI22/00338) holds a contract funded by the Carlos III Health Institute. Eliseo Albert (Juan Rodés Contract; JR20/00011) holds a contract funded by the Carlos III Health Institute (co-financed by the European Regional Development Fund, ERDF/FEDER).