According to WHO and European Centre for Disease Control, only thirteen imported cases of cholera have been notified by Spain in the last 15 years, the last one in 2013. On August 20, 2015, Vibrio cholerae was isolated from two immunocompetent female patients (47 and 62 years-old) returning from Dominican Republic (DR). No patient had previously received the cholera vaccine. Both patients showed acute and watery diarrhoea (12–15stools/day, no blood and mucus), nausea, abdominal pain, and malaise. They had returned from a tourism trip to Bonao (DR) four days before, where they had stayed for 15 days. Physical examinations were normal, except for lightly dehydrated mucoses and skin. Laboratory analysis revealed hypokalemia and altered C-reactive protein. The patients informed the physician that a cholera outbreak had recently been detected in Bonao. This was related to drinking contaminated water from Masipedro river. They had not drunk contaminated water (only bottled water) or drinks with ice cubes, but they had swum in the Masipedro river during their stay. This information was notified as suspected cholera cases to the public health authorities. A blood and faecal culture from each patient were sent to the microbiology department. Intravenous fluids, metoclopramide and antibiotic treatment (Doxycycline 100mg for 3 days) were started. They remained afebrile and this led to quick recovery from their gastroenteritis.
Because of the epidemiological background, testing for V. cholerae was included [enriched alkaline peptone water (1% NaCl pH 8.5) and selective thiosulfate citrate bile salts sucrose (TCBS) agar (Becton–Dickinson)]. The TCBS showed yellow colonies (sucrose fermenting) which were identified as V. cholerae by API20E® (99.0%, Bionumber: 5346120; bioMérieux) and MALDI-TOF MS (score 1.53; Brucker Daltonics). The isolates reacted with polyvalent anti-O1 antisera (BD Difco™ V. cholerae antisera, Becton–Dickinson). They were resistant to the O/129 vibriostatic agent 150μg (Thermo Scientific Oxoid). The isolates were confirmed as V. cholerae O1 biotype El Tor serotype Ogawa in the National Centre for Microbiology. Moreover, the presence of virulence determinants and toxin coding genes were detected by PCR.1 Pulsed-field gel electrophoresis2 and MLVA3 showed the same pulsotype and pattern in both strains (Table 1).
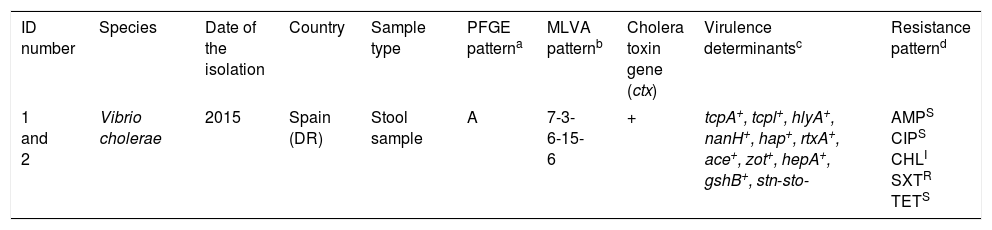
Characteristics of Vibrio cholerae O1 El Tor Ogawa strains isolated from two patients returning from Dominican Republic.
| ID number | Species | Date of the isolation | Country | Sample type | PFGE patterna | MLVA patternb | Cholera toxin gene (ctx) | Virulence determinantsc | Resistance patternd |
|---|---|---|---|---|---|---|---|---|---|
| 1 and 2 | Vibrio cholerae | 2015 | Spain (DR) | Stool sample | A | 7-3-6-15-6 | + | tcpA+, tcpl+, hlyA+, nanH+, hap+, rtxA+, ace+, zot+, hepA+, gshB+, stn-sto- | AMPS CIPS CHLI SXTR TETS |
Multi-locus variable number tandem repeat analysis types were assigned by combining the number of repeat units of each locus in the order VC0147, VC0436-7, VC1650, VCA0171 and VCA0283.
tcpA, classical biotype and tcpl, the toxin-coregulated-pilus associated genes; hlyA, hemolysin gene (classical biotype); nanH, neuraminidase gene, hap, hemagglutinin protease gene; rtxA, cytotoxic actin cross-linking repeats in toxin gene; ace, accessory cholera enterotoxin; zot, zonula occludens toxin; hepA, the transcriptional regulator; gshB, glutathione synthetase; stn-sto, heat-stable toxin.
An antimicrobial susceptibility test was performed for each isolate using the disc diffusion method (Becton–Dickinson) on Mueller–Hinton agar, and interpreted according to Clinical and Laboratory Standards Institute guidelines (M45 3rd edition). AMP (10μg): Ampicillin, (18mm); CIP (5μg): Ciprofloxacin, (25mm); CHL (30μg): Chloramphenicol (13mm); SXT (1.25/23.75μg): Trimethoprim/sulfamethoxazole (6mm), TET (30μg): Tetracycline, (25mm).
In locations such as Europe where cholera is a sporadic illness, the clinical microbiology laboratory should be informed of any clinical and epidemiological suspicion of cholera (e.g.: traveller's patient history, risk activities, watery diarrhoea or rice water, current cholera outbreaks). The culture, biochemical identification, commercial identification systems, and immunoassay are time consuming and often lack sensitivity and specificity due to high phenotypic diversity. Furthermore, there have been detected strains resistant to vibriostatic compound O/129.4 All Vibrio species other than V. cholerae have been successfully identified by MALDI-TOF MS system, but this tool has failed to identify V. cholerae isolates to the species-level, genus-level, or even not to get a reliable identification.5 However, a newly generated established MALDI-TOF MS database, the VibrioBase, has been very useful for rapid identification of human-pathogenic Vibrio spp.6
The genes that code for the virulence factors (ctx and tcpA) traditionally associated with epidemic V. cholerae O1 and O139 were detected. Both isolates presented the same pattern of virulence factors, and there were identified genes such as hlyA or rtxA in agreement with studies from other countries.4,7 Extensive genetic variation among cholera strains has been observed by MLVA from different regions and times of collection.8 Based on the history of travel to Bonao, it is reasonable to assume the patient isolates were related to the outbreak strain. On the other hand, the high negative predictive value of the molecular multiplex gastrointestinal panels has led to the suggestion that they be used as screening tools especially in outbreaks since it would improve diagnosis of infectious gastroenteritis, decisions regarding patient isolation and reduce nosocomial transmission.9
There have been reports of increased drug resistance towards antibiotics commonly used among the strains of V. cholerae, causing serious problems in management of cholera cases mainly in developing countries. These have appeared in many cholera endemic countries, including ampicillin [resistance (R): 0–100%], tetracycline (0–81.8%), ciprofloxacin (0–9.1%), chloramphenicol (3.2–9.1%), or cotrimoxazole (83.3–100%).10 Our cases underline the importance of testing for V. cholerae in potentially exposed patients with acute diarrhoea who return from epidemic areas. Continuous circulation inside and outside of some American, African or Asian countries increases the likelihood of imported cases. Travellers must be informed about the potential risks of cholera, symptoms, and precautions to prevent disease and secondary transmission. Conventional culture media, MALDI-TOF and multiplex gastrointestinal PCR help us to establish an adequate diagnosis.
FundingNone declared.
Conflict of interestNone declared.
The authors thank to Jesús Castilla Catalán of the Public Health Institute of Navarre and Xabier Beristain Rementería of the Complejo Hospitalario de Navarra for reviewing the manuscript.