Campylobacter spp. is a major cause of acute bacterial diarrhea in humans worldwide, and C. coli is responsible for 10% of the cases.
Materials and methodsA study was made of the antimicrobial susceptibility using the E-test®, and the clonal relationship using PCR-RFLP, of the flaA gene, as well as PFGE techniques on 43 C. coli clinical isolates.
ResultsOnly 49% and 2% of the isolates were susceptible to erythromycin and ciprofloxacin, respectively. Imipenem and clindamyicn, with 100% and 84% of the strains, respectively, being susceptible, were the most active antimicrobials. The PCR-RFLP of flaA gene technique grouped fourteen isolates into six clusters, while the PFGE technique grouped eleven isolates into five clusters.
ConclusionCiprofloxacin and erythromycin are not suitable for the treatment of C. coli infections. Clindamycin could be considered as a therapeutic alternative in cases of enteritis, while imipenem is the best alternative for extra-intestinal infections. Both PFGE and PCR-RFLP can be useful to detect clones.
Campylobacter spp. es la principal causa de diarrea bacteriana en el mundo, siendo. C. coli responsable del 10% de los casos.
Material y métodosHemos estudiado la sensibilidad antibiótica mediante E-test® y la relación clonal por PCR-RFLP del gen flaA y ECP de 43 cepas de C. coli.
ResultadosSolo el 49 y el 2% de todas las cepas estudiadas fueron sensibles a eritromicina y a ciprofloxacino, respectivamente. El imipenem, con el 100% de cepas sensibles, y la clindamicina, con el 84%, fueron los antibióticos más activos. La técnica de PCR-RFLP del gen flaA agrupó13 cepas en 6 clones, y la ECP, 11 cepas en 5 clones.
ConclusionesCiprofloxacino y eritromicina no son antimicrobianos adecuados para tratar las infecciones por C. coli. La clindamicina se puede considerar como una alternativa terapéutica en caso de enteritis, mientras que el imipenem es la mejor alternativa en la infección extraintestinal. Tanto la ECP como la PCR-RFLP del gen flaA son técnicas útiles para detectar clones.
Campylobacter spp. is a major cause of acute bacterial diarrhea in humans worldwide, especially in developed countries. Campylobacter jejuni is responsible for a majority of cases (about 90% of cases 10% is associated to C. coli. The most important sources of human infection are the handling and consumption of contaminated meat from domesticated animals, but other potential sources are described.1 A large proportion of cases are considered as sporadic disease, although outbreaks of campylobacteriosis have been identified.2
The antimicrobials resistance rates among Campylobacter spp. have increased overtime, and numerous studies have been performed to understand the mechanisms of antibiotic resistance,3,4 its implication in the fitness of the bacteria5 as well as the epidemiology of this antimicrobial resistance.6 Several genotyping methods have been developed to identify as well as to study the epidemiology of Campylobacter spp., which include among others pulsed-field gel electrophoresis (PFGE) and flaA gene typing.7,8 The aim of this study is to determine the susceptibility of C. coli clinical isolates to several antimicrobials. Additionally, two molecular methods, PCR-RFLP of flaA gene and pulsed-field gel electrophoresis (PFGE), were used to establish the clonal relationship of the isolates.
Materials and methodsStrainsFrom January 2011 to July 2012, 406 Campylobacter spp. (346 C. jejuni and 60 C. coli) isolates were obtained from 13.966 human fecal samples, submitted to the clinical microbiology laboratory of the University Hospital Virgen del Rocío in Seville, Spain. The strains were isolated from fecal samples streaked onto modified Skirrow medium agar after 48h of micro-aerobically incubation at 42°C. Identification of C. coli included Gram staining, oxidase and catalase activities, the hippurate test and an additional method, the matrix assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF).9
Antimicrobial susceptibility testingAmong the 60 C coli isolates, 10 were duplicated isolates and 7 could not be recovered. The susceptibility testing to erythromycin, ciprofloxacin, tetracycline, tobramycin, amoxicillin–clavulanic, clindamycin and imipenem of the 43 C. coli isolates was studied by the E-test® (Biomerieux) method. The MIC breakpoints for erythromycin, ciprofloxacin, tetracycline, tobramycin and amoxicillin–clavulanic established by the FSM (French Society for Microbiology) were used,10 and for imipenem and clyndamycin those of the non-related species EUCAST breakpoints and the epidemiological of EUCAST breakpoints11 respectively were used for clinical categorization of the isolates.
Statistical analysis with Yates test, Chi2 and Fisher's Exact Probability Test was performed as needed.
Molecular genotypingForty-three isolates were analyzed by Restriction fragment length polymorphism of the flaA gene (PCR-RFLP flaA) with DdeI enzyme as previously described,7 and by PFGE using the standardized PulseNet protocol, with the Salmonella Braenderup H9812 strain restricted with XbaI as a size standard.8 In both techniques, profiles were analyzed and compared using FQuest® (Biorad) program, and clones were defined as isolates with indistinguishable banding patterns.
ResultsA male predominance (60% of cases) of two age groups was found among the patients with C. coli infections: children aged 1–5 years (38%) and adults over 40 years old (21.4%). 83.8% of hospitalized patients belonged to the latter group, and all of them were suffering from severe underlying diseases like solid organ transplantation and chronic kidney disease.
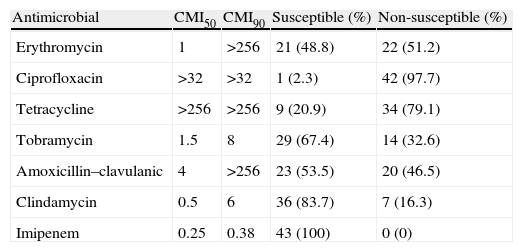
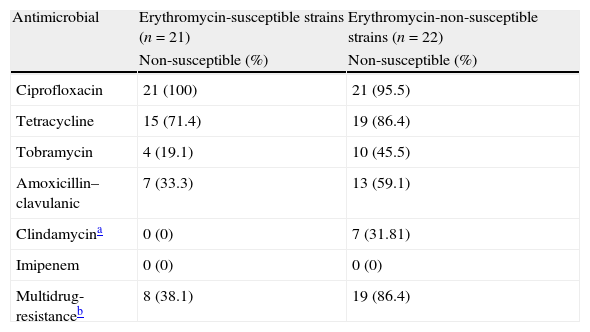
Although erythromycin and ciprofloxacin are considered the treatments of choice, only 21 (49%) and 1 (2%) isolates were susceptible to them respectively. Among the remaining antibiotics tested, the most active were imipenem and clindamycin, with 43 (100%) and 36 (84%) susceptible strains (Table 1). A higher rate of antimicrobial non-susceptibility was found among the erythromycin non-susceptible strains. In this group, imipenem is the only active antimicrobial against all these strains (Table 2). Additionally, in this group we found a 86% of multi-resistance rate in comparison with the 38% found in the erythromycin- susceptible strains group.
Antimicrobial susceptibility of the 43 strains of Campylobacter coli.
| Antimicrobial | CMI50 | CMI90 | Susceptible (%) | Non-susceptible (%) |
| Erythromycin | 1 | >256 | 21 (48.8) | 22 (51.2) |
| Ciprofloxacin | >32 | >32 | 1 (2.3) | 42 (97.7) |
| Tetracycline | >256 | >256 | 9 (20.9) | 34 (79.1) |
| Tobramycin | 1.5 | 8 | 29 (67.4) | 14 (32.6) |
| Amoxicillin–clavulanic | 4 | >256 | 23 (53.5) | 20 (46.5) |
| Clindamycin | 0.5 | 6 | 36 (83.7) | 7 (16.3) |
| Imipenem | 0.25 | 0.38 | 43 (100) | 0 (0) |
Antimicrobial resistance among erythromycin susceptible and non-susceptible Campylobacter coli strains.
| Antimicrobial | Erythromycin-susceptible strains (n=21) | Erythromycin-non-susceptible strains (n=22) |
| Non-susceptible (%) | Non-susceptible (%) | |
| Ciprofloxacin | 21 (100) | 21 (95.5) |
| Tetracycline | 15 (71.4) | 19 (86.4) |
| Tobramycin | 4 (19.1) | 10 (45.5) |
| Amoxicillin–clavulanic | 7 (33.3) | 13 (59.1) |
| Clindamycina | 0 (0) | 7 (31.81) |
| Imipenem | 0 (0) | 0 (0) |
| Multidrug-resistanceb | 8 (38.1) | 19 (86.4) |
36 band patterns (mean number of 4–7 bands) were found with the PCR-RFLP of flaA gene technique and only thirteen isolates grouped in six clusters. With the PFGE technique 37 band patterns (mean number of 8 bands) and eleven isolates were grouped in five clusters. Combining both techniques, we identified 3 clones, two clones with two strains and one with three strains, among all strains studied. The first clone included two strains isolated from the same patient with 4 months of difference, suggesting a reinfection or a recurrence. The second clone included three strains, two of them isolated from two brothers with simultaneous diarrhea syndrome. The last clone included two isolates from patients with no evident spatial nor temporal relationship.
DiscussionOur results are similar to those described by Olson et al.,12 who found a 20% higher incidence of C. coli infection in the male population, and in children aged 1–4 years and over 40 years. In developing countries, where exposure to the bacteria is very high, there is a peak in childhood which then decays strongly due to the protective antibodies.1 However, in developed countries, as in this study, with less exposure to the organism two peaks are found, one at childhood and the second in middle-aged adult as we have found.
Erythromycin is considered the antimicrobial of choice for Campylobacter infections treatment, but a high level of macrolides resistance in C. coli have been described, and we have found a 51% of non-susceptible C. coli human isolates. Additionally, several studies have warned of the increasing incidence of the resistance rate to this antimicrobial in various parts of the world, with especially high rates in Spain.3 In our study, ciprofloxacin, with 98% of resistant strains, is a highly ineffective treatment against infections.
Although antimicrobial treatment is not needed in most cases of C. coli gastrointestinal infections; in young children, the elderly, pregnant women, immunocompromised patients, and in serious, prolonged or extraintestinal infections, it requires antimicrobial treatment. In these cases, alternative antibiotics should be used in the treatment of infections due to resistant strains.
According to our results, clindamycin (84% of susceptible strains) could be considered as a therapeutic alternative in cases of enteritis although in other studies in both clinical and animal strains variable rates of resistance are found.13 The best alternative for extraintestinal infections C. coli is imipenem, which is active against all the clinical isolates, as previously suggested by other authors.13,14
In the case of aminoglycosides, tobramycin could also be a good alternative for extraintestinal infections,13 but its use should be guided by the results of susceptibility testing since we found a 33% of non-susceptible strains. Although in most countries the resistance to aminoglycosides is scarce, in others like China resistance rates of 20% have been found.4
Furthermore, we have shown a very high percentage of multiresistant strains among the erythromycin non-susceptible strains, which will increase the challenge of the treatment of these infections, with imipenem being the only alternative therapeutic in severe clinical situations.
Both PFGE and PCR-RFLP of the flaA gene allow clonal differentiation of the strains studied. The PFGE technique generates an adequate number of bands which gives greater power of discrimination. We obtained a high percentage of unique strains: 74% and 69% by PFGE and PCR-RFLP flaA gene respectively. This genetic heterogeneity is also found in numerous studies of human, animal and environmental isolates.15,16
In those cases where an outbreak is suspected both genotypic techniques can be useful to detect clones and could help to understand the epidemiology of these infections.
Conflict of interestThe authors declare no conflict of interest.
Part of this work has been presented in the XVII SEIMC Congress, celebrated in Zaragoza, Spain, in 2013.