Collateral damage caused by antibiotic use includes resistance, which could be reduced if the global inappropriate use of antibiotics, especially in low-income countries, could be prevented. Surveillance of antimicrobial consumption can identify and target practice areas for quality improvement, both in the community and in healthcare institutions. The defined daily dose, the usual adult dose of an antimicrobial for treating one patient for one day, has been considered useful for measuring antimicrobial prescribing trends within a hospital. Various denominators from hospital activity including beds, admissions and discharges have been used to obtain some standard ratios for comparing antibiotic consumption between hospitals and countries. Laboratory information systems in Clinical Microbiology Services are the primary resource for preparing cumulative reports on susceptibility testing results. This information is useful for planning empirical treatment and for adopting infection control measures. Among the supranational initiatives on resistance surveillance, the EARS-Net provides information about trends on antimicrobial resistance in Europe. Resistance is the consequence of the selective pressure of antibiotics, although in some cases these agents also promote resistance by favouring the emergence of mutations that are subsequently selected. Multiple studies have shown a relationship between antimicrobial use and emergence or resistance. While in some cases a decrease in antibiotic use was associated with a reduction in resistance rates, in many other situations this has not been the case, due to co-resistance and/or the low biological cost of the resistance mechanisms involved. New antimicrobial agents are urgently needed, which coupled with infection control measures will help to control the current problem of antimicrobial resistance.
El daño colateral más importante derivado del uso de los antibióticos es la aparición de resistencias bacterianas. La prescripción inadecuada de los antibióticos está íntimamente relacionada con este efecto, observado globalmente a nivel mundial, pero principalmente en países con recursos económicos limitados. La estrecha vigilancia del consumo de los antibióticos puede ser de gran ayuda para identificar cuáles son los problemas relacionados con la prescripción de estos fármacos e introducir las estrategias necesarias para evitarlos, tanto en el ámbito ambulatorio como en el hospitalario. La dosis diaria definida, referida a la dosis usual de un antimicrobiano concreto, destinada al tratamiento diario de un paciente, se ha considerado útil para el estudio de las tendencias de consumo de los antibióticos en el hospital. Esta unidad se ha introducido en diversas fórmulas que incluyen diversos denominadores correspondientes a la actividad hospita laria, entre ellos el número de camas, ingresos y altas. Todo ello, con el objetivo de obtener una serie de indicadores estandarizados que se utilizan para efectuar comparaciones sobre el uso de antibióticos entre distintos hospitales y países. Los sistemas de información del laboratorio son las fuentes primarias de datos para la preparación de informes acumulados de sensibilidad. Esta información es útil para planificar tratamientos empíricos y adoptar medidas de control de infección. Entre las iniciativas supranacionales de vigilancia de la resistencia, la red EARS-Net proporciona información acerca de las tendencias de resistencia en Europa. La resistencia es consecuencia de la presión selectiva de los antimicrobianos, aunque en ocasiones estos agentes también promueven la resistencia al favorecer la aparición de mutaciones seleccionadas posteriormente. Múltiples estudios indican la relación entre el uso de antimicrobianos y la aparición de resistencias. Aunque en algunos casos una disminución del uso de un antimicrobiano se asocia a una reducción en las tasas de resistencia a este, en muchas otras situaciones no sucede así, debido a la corresistencia o al bajo coste biológico del mecanismo implicado. Son necesarios nuevos antimicrobianos, que junto con medidas de control de infección ayudarán a paliar el problema de la resistencia.
Antibiotics have increased life expectancy. Self-medication occurs in many countries where antibiotics are classified as prescription-only medicines. Currently, microbial resistance to treatment with antibiotics constitutes an important public health problem, especially in the hospital environment. In this environment we find significant complexity and density of antibiotic use.
Surveillance of antimicrobial resistance can identify trends in resistance patterns and novel resistances. Antimicrobial stewardship initiatives and infection control programmes play an important role in decreasing inappropriate use and halting the dissemination of resistance. Education of professionals and the public should focus on changing behaviour rather than exclusively increasing knowledge, as the latter could have a paradoxical effect by increasing demand and prescription. Behaviour change should target all prescribers, including veterinarians, since microbes know no boundaries between animals and humans and are capable of exchanging resistance genes.1 This section is focused on certain aspects related to measures of antimicrobial consumption and bacterial resistance.
Ways of measuring antimicrobial consumptionA European Commission press release dated November 17, 2011 outlined an action plan against bacterial resistance to last-line antibiotics, comprising 12 specific measures to be implemented in the next five years.2 Two of the measures aim to heighten awareness regarding the appropriate use of antimicrobials and to strengthen surveillance systems of bacterial resistance and antimicrobial consumption in medicine.
In order to promote rational use and to avoid the development of resistance, antimicrobial consumption3 in groups of hospitals and departments must be measured and compared. Trends in antibiotic consumption must also be monitored. For this to be possible, the data must be expressed in the same units of measurement. At present, comparisons with other hospitals or countries are not always possible, as different bibliographic resources may contain different measurement units—a situation that generates a certain degree of confusion.4–8
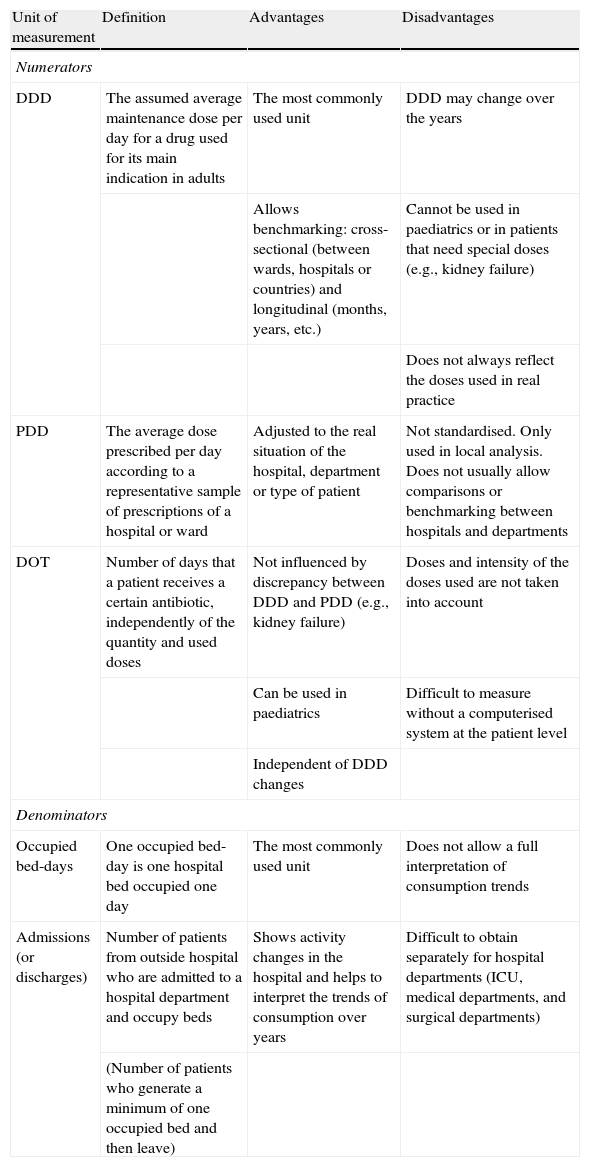
Usually, consumption is expressed as a quotient consisting of a numerator and a denominator. Today, the three measurement units most often used as the numerator are:9 DDD10 (defined daily doses), PDD6 (prescribed daily doses) and DOT11 (days of therapy, a new unit used in the US). The definitions and the advantages and disadvantages of each unit of measurement are specified in Table 1.
Units of measurement most commonly used to calculate antimicrobial consumption
| Unit of measurement | Definition | Advantages | Disadvantages |
| Numerators | |||
| DDD | The assumed average maintenance dose per day for a drug used for its main indication in adults | The most commonly used unit | DDD may change over the years |
| Allows benchmarking: cross-sectional (between wards, hospitals or countries) and longitudinal (months, years, etc.) | Cannot be used in paediatrics or in patients that need special doses (e.g., kidney failure) | ||
| Does not always reflect the doses used in real practice | |||
| PDD | The average dose prescribed per day according to a representative sample of prescriptions of a hospital or ward | Adjusted to the real situation of the hospital, department or type of patient | Not standardised. Only used in local analysis. Does not usually allow comparisons or benchmarking between hospitals and departments |
| DOT | Number of days that a patient receives a certain antibiotic, independently of the quantity and used doses | Not influenced by discrepancy between DDD and PDD (e.g., kidney failure) | Doses and intensity of the doses used are not taken into account |
| Can be used in paediatrics | Difficult to measure without a computerised system at the patient level | ||
| Independent of DDD changes | |||
| Denominators | |||
| Occupied bed-days | One occupied bed-day is one hospital bed occupied one day | The most commonly used unit | Does not allow a full interpretation of consumption trends |
| Admissions (or discharges) | Number of patients from outside hospital who are admitted to a hospital department and occupy beds | Shows activity changes in the hospital and helps to interpret the trends of consumption over years | Difficult to obtain separately for hospital departments (ICU, medical departments, and surgical departments) |
| (Number of patients who generate a minimum of one occupied bed and then leave) | |||
DDD: defined daily doses; DOT: days of therapy; PDD: prescribed daily doses.
Each year, the WHO's International Working Group for Drug Statistic Methodology of Norway establishes the DDD for each drug and administration route.
At the hospital level, the most frequently used denominators are: 100 (or 1,000) occupied bed-days (OBD) and 100 admissions (or discharges).12 The calculation of DDD/100 OBD is carried out according to the formula: DDD/100 OBD = consumption/DDD × 100/OBD, where the consumption and the DDD are expressed in the same units (grams).
At the primary care level, the most frequently used unit is DDD/1,000 inhabitants/day.
The monitoring of antimicrobial consumption is usually presented in a report9 that records: a) The periodicity of measurement (e.g., annual, six-month, quarterly, monthly, before and after an intervention); b) The department studied (overall, medical or surgical department, ICU, a certain ward); c) The clinical indications (e.g., antipseudomonals); d) The level of data aggregation (e.g., therapeutic group or subgroup, drug, medicine).
The reports may be cross-sectional (a certain year or month) or longitudinal, in order to track the evolution of consumption over time.
For a correct evaluation of trends of consumption in hospitals over time, the variations in the hospital indicators and consumption must be expressed in DDD/100 OBD and DDD/100 discharges.12
Information about trends of hospital consumption of antimicrobials in Spain is scarce. However, in the autonomous community of Catalonia the VINCat monitoring program compiled data from 54 hospitals on antibacterial and antifungal consumption in the period 2007–2011.13–16
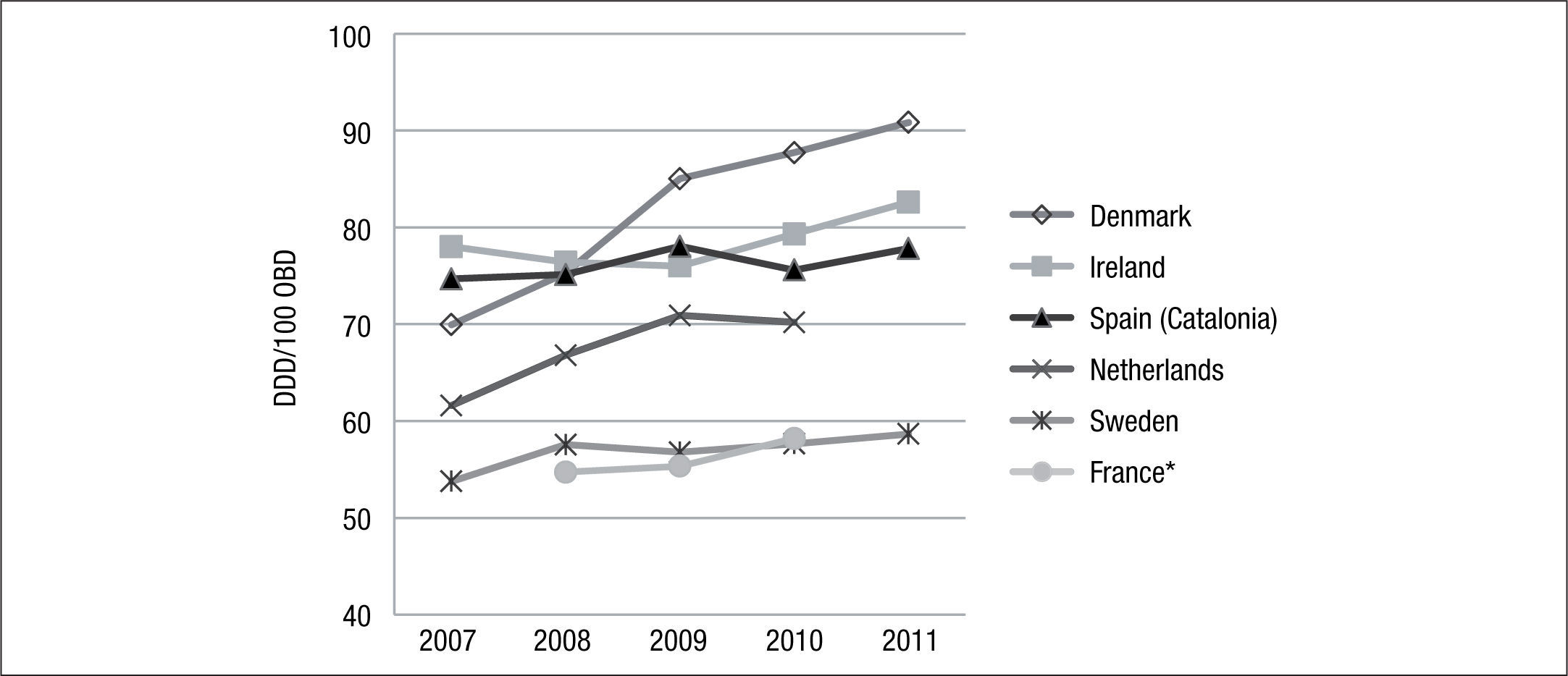
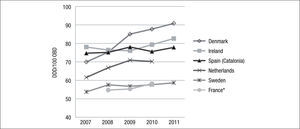
In Europe, several countries have carried out studies of antibacterial consumption over time. Examples include Denmark (DANMAP17), Netherlands (NETHMAP18–19), France (CCLIN20–23), Sweden (SWEDRES24,25) and Ireland (HSE-HPSC26) (Fig. 1). In general, antibiotic consumption presents an upward trend in all these countries.
Evolution of human consumption of antimicrobial agentsAlthough most antimicrobials are prescribed in the community,27 microorganisms isolated from hospital infections usually show more resistant profiles than microorganisms from community infections,28 due to the fact that the proportion of patients receiving antimicrobial agents is much higher in hospitals than in the community,29 and for this reason the exerted selective pressure is much higher in hospitals.
In order to see the evolution of antibiotic consumption in the European Union, we will analyse the data published by the ESAC Net. The European Surveillance of Antimicrobial Consumption (ESAC) network is an international data collection network that aims to improve antimicrobial prescribing by collecting data on patterns of antibiotic prescribing utilizing a standard validated method.30 ESAC-Net is today a Europe-wide network of national surveillance systems that provides independent reference data on antimicrobial consumption in Europe, reported by 29 EU/EEA countries. It collects and analyses data from the community (primary care) and the hospital sector.
Performing a methodical analysis of the evolution of antibiotic consumption at European level is complex, due to the fact that published data have not been obtained in a homogeneous way. Data on antimicrobial consumption in the community were obtained from the Ministry of Health or the national medicine agency of half of the countries. About one-third of the countries reported reimbursement data, while the remaining countries reported sales data. Three countries reported both sales and reimbursement data. For most countries, the data coverage was reported as being 100%. Germany, Luxembourg, the Netherlands and Portugal reported data that covered 80–95% of the population. Most countries provided data on all antimicrobial categories under surveillance by ESAC-Net. Ireland, Poland, Spain and the United Kingdom only reported data on antibacterials for systemic use (ATC group J01).31
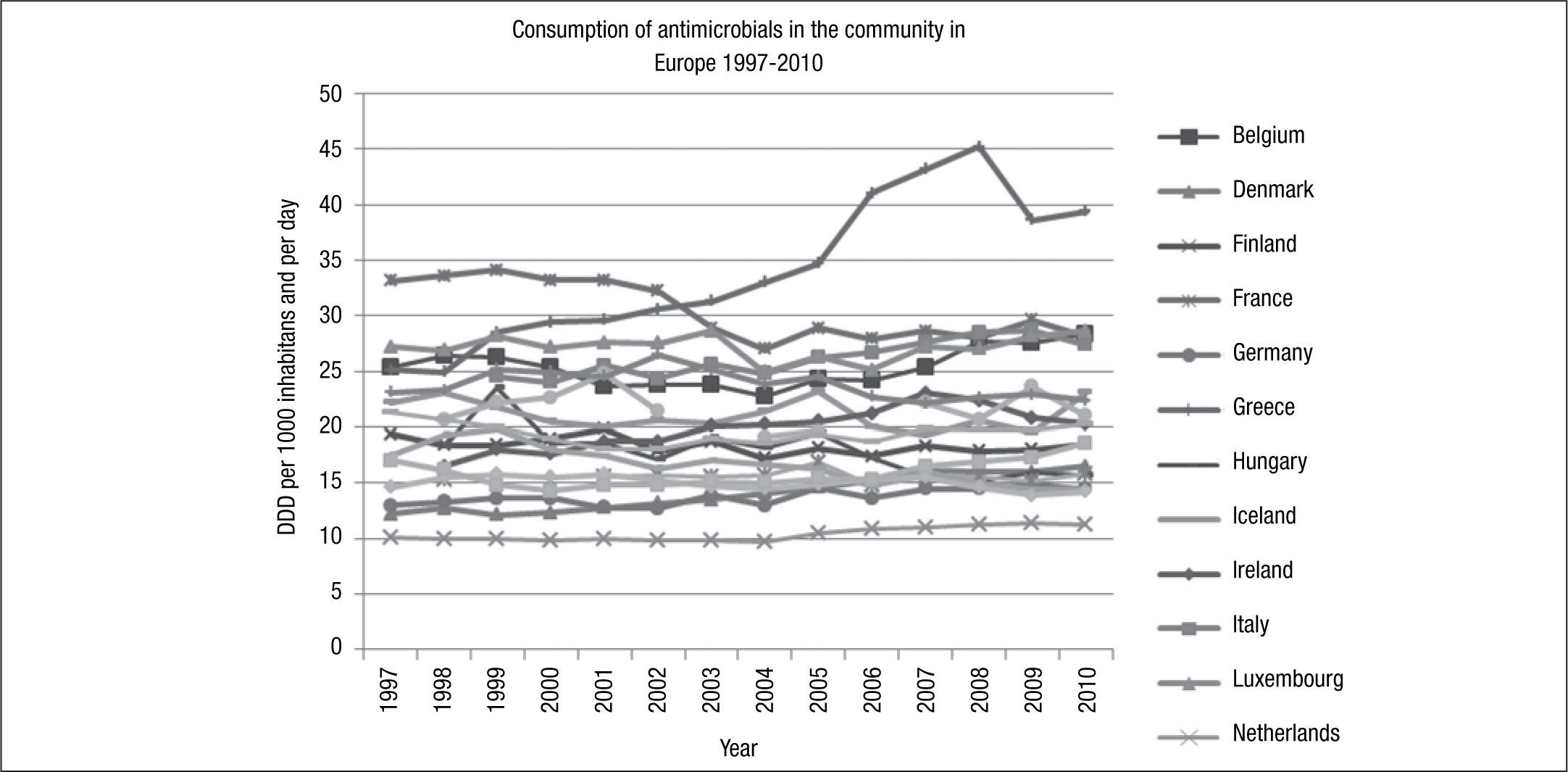
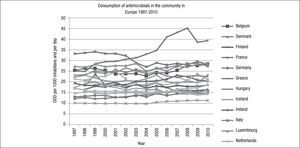
The variation in antibiotic consumption has remained higher than 3 since 1997, and France was the country with the highest consumption (33.1 DDD/1,000 inhabitants/day) and the Netherlands with the lowest consumption (10.1 DDD/1,000 inhabitants/day) until 2010, at which point Greece became the country with the highest consumption (39.4 DDD/1,000 inhabitants/day), and Estonia the country with the lowest consumption (11.1DDD/1,000 inhabitants/day) (Fig. 2).
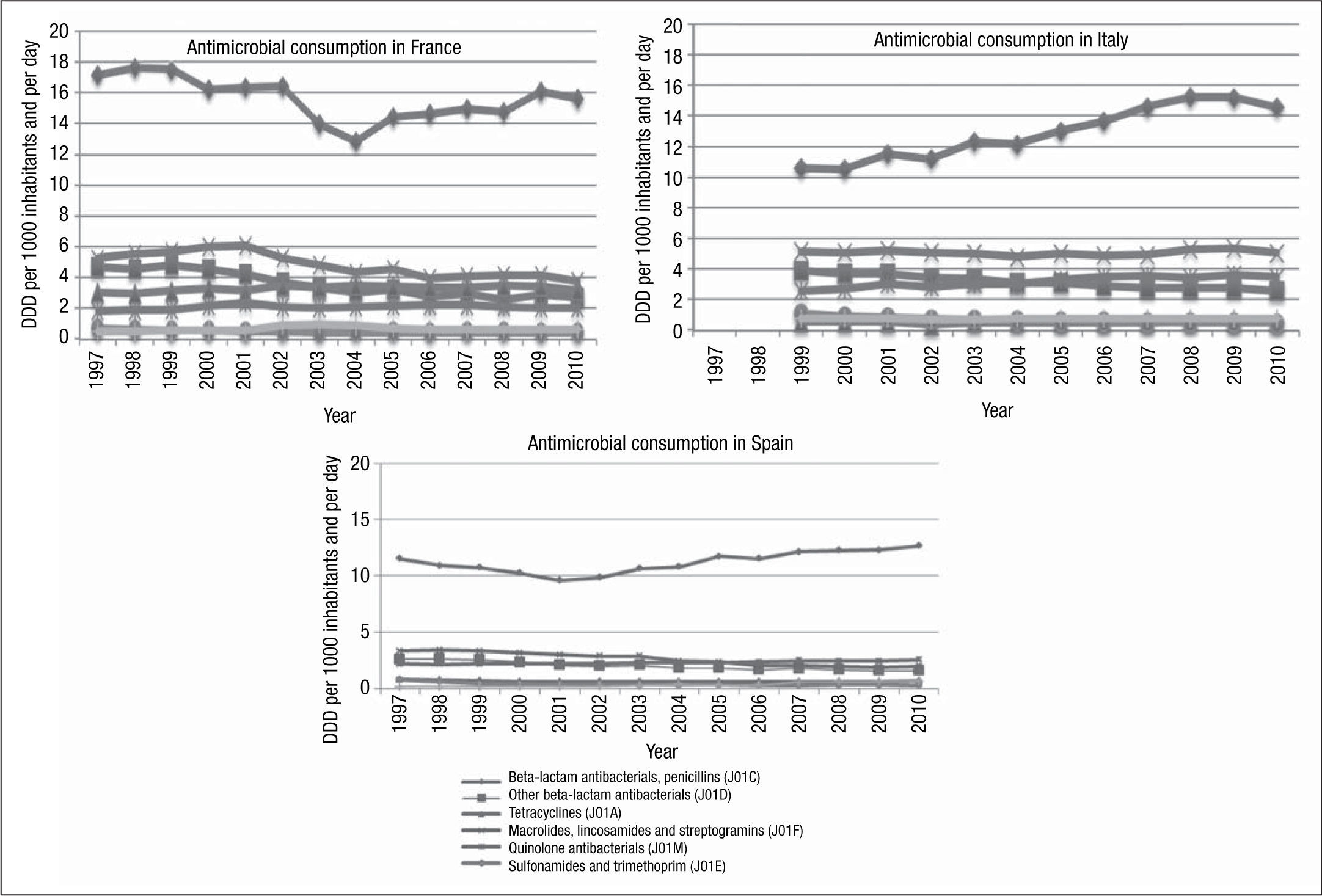
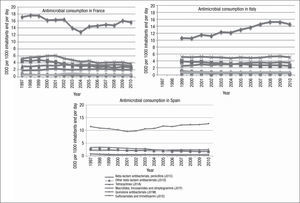
During the study period, a north-south gradient has been observed, with the lowest consumption (<16.7 DDD/1,000 inhabitants/day) in the north of Europe, e.g., Scandinavian and Baltic countries, and the highest consumption (≥22.4 DDD/1,000 inhabitants/day) in the south of Europe, e.g., Greece, Italy and Portugal.31 France is in the group of countries with a range of consumption between 28 and 33 DDD/1,000 inhabitants/day, in 2010. As with other countries, in France, there was a higher consumption of penicillins (J01C) and tetracyclines (J01A) (Fig. 3). Italy, for example, in 2010, belonged to the group of countries of intermediate consumption, and an important increase in consumption of penicillins (J01C) can be appreciated during recent years. However, in Spain, only a moderate increase in consumption of penicillins was reported (Fig. 3).
In 2010, the median consumption of antibacterials for systemic use (ATC group J01) was at the same level as in 2009, whereas a generally decreasing trend has been observed from 1999 to 2004 followed by a gradual increase from 2004 to 2008.
Measuring bacterial resistanceData to measure resistance to antimicrobial agents are first obtained at Clinical Microbiology services, based on cumulative tabulated susceptibility testing results of isolates from individual patients. In Spain and many other countries, this is most often achieved using the laboratory information system (LIS), or susceptibility testing system software. The free Windows-based WHONET software (http://www.who.int/drugresistance/whonetsoftware/en) can also interact with susceptibility test instruments using BackLink. SaTScan, another public software programme, can define statistically significant clusters or organisms that might be causing an outbreak.32 After the report on susceptibility testing with data from the LIS (or other system) is completed, it is recommendable to perform a careful analysis of the information obtained, to discard possible errors, and to confirm “impossible” or infrequent results.
Monitoring resistance to antimicrobial agents is relevant at local, regional-national and supranational levels.33,34 Local data are important for guiding empirical therapy and for identifying concrete problems of resistance that need infection control measures.
The CLSI has approved a document (M39-A3) aimed at developing guidelines for routine generation, storage and compilation of susceptibility data and for providing indications about effective use of cumulative susceptibility statistics.35 Data should be presented at least annually, considering organisms cultured from diagnostic samples (but not from surveillance studies) and drugs that are normally tested. It has been recommended that only species for which 30 or more isolates have been tested be reported. Periods longer than one year or data for more than one facility in the same area can be considered for reaching the indicated 30 isolates, whenever this is clearly stated in a footnote of the report. If for some drug(s) only selected isolates are reported, this should also be indicated, to avoid a biased interpretation of the data.
When total data from the laboratory database are considered, an overestimation of resistance frequently occurs, particularly because microbiological studies are more likely performed on patients in whom the therapeutic response is not good (which may be partly related to bacterial resistance). For this reason, it is recommended that the local report be based on the first isolate per patient obtained in the considered period, with independence that a more resistant isolate may appear as a consequence of a complicated infection or if several samples with the same organism are obtained from the same patient. Various approaches can be considered to eliminate repeated isolates, but the most common approach is based on the definition of a minimal period for which isolates of the same species represent non-duplicate ones.36–38 However, It is also important to consider that when duplicates are removed from the final report, the impact of the development of resistance during therapy (an obviously relevant clinical event) is not appropriately reflected.
Cumulative reports normally include data on percentages of susceptible isolates, as most clinicians do not treat infections caused by organisms with a clinical category of intermediate. Relevant exceptions to this general rule include presenting data on Streptococcus pneumoniae or Streptococcus group viridans with intermediate susceptibility to penicillin. On the other hand, clinical microbiologists may find it more interesting to report percentages of resistant (or non-susceptible) isolates, as they are helpful in the follow-up of resistance trends.
Many laboratories present detailed information for selected assistance units (e.g., the ICU), samples (e.g., blood cultures or urine cultures) or patient populations, for a particular clinical purpose. It is also common for data on particular resistant phenotypes (e.g., enterobacteria producing extended-spectrum beta-lactamase, carbapenemase-producing organisms, methicillin-resistant Staphylococcus aureus) to be included. An important aspect is making the available information accessible to clinicians and other relevant players (e.g., infection control committees, management, and public health authorities). “Pocket guides” and internet tools with personalized access are helpful in this regard.
There is increasing interest among public health authorities and other entities in obtaining regional or national information about resistance trends. This information can be obtained by integrating data from multiple laboratories into a single database. Research groups and scientific societies are also promoting studies on resistance trends, frequently also looking for precise information on concrete mechanisms of resistance and their clinical impact.
Several initiatives have also been organised at an international level for surveillance of resistant bacteria.39 Multiple problems hamper the organisation of international databases for the evaluation of antimicrobial resistance, particularly those related to the standardisation of the susceptibility testing assays and the definition of breakpoints for clinical categorisation. The European Antimicrobial Resistance Surveillance Network (EARS-Net, http://ecdc.europa.eu/en/activities/surveillance/EARSNet/database/Pages/database.aspx) is probably the most important public database in the field. In addition, many private sources (usually pharmaceutical companies) are sponsoring surveillance resistance programs focused on particular drugs, group of organisms and/or defined types of infections. EARS-Net collects information from some 900 laboratories (in approximately 1400 hospitals caring for about 100 million people). Multiple national networks collect data in their own countries and send this information to the central database at European Centre for Disease Prevention and Control (ECDC, http://www.ecdc.europa.eu/en/Pages/home.aspx). EARS-Net contains data on seven bacterial species (S. pneumoniae, S. aureus, E. faecalis, E. faecium, E. coli, K. pneumoniae and P. aeruginosa) cultured from invasive infections, and presents comparative information on antimicrobial resistance over time for multiple European countries. Since 1999, more than 400,000 invasive isolates have been included in its interactive database and annual reports.
Antibiotic pressure and the development of bacterial resistanceIt is currently accepted that exposure to antibiotics does not directly determine the appearance of resistance in bacteria; it instead causes selective pressure on susceptible bacteria, allowing the proliferation of resistant organisms from pre-existing resistant subpopulations. Antibiotics should be considered, in general terms, as selective agents in a “natural” (Darwinian) process of survival of the fittest. Mutants may appear spontaneously because of mutations generated during DNA replication or after horizontal acquisition of resistance genes from other bacteria.
Bacteria may contain “pre-resistance” genes (related to housekeeping activities), which can evolve into actual resistance genes by gene duplication, a rather frequent process in which one of the genes maintain its original function and the other include mutation(s) conferring resistance properties. These genes usually confer a low level of resistance, but it allows the bacteria to survive in small concentrations of antibiotics.40
Within a considered population, some bacteria (persisters) can survive the lethal action of antibiotics.41 They behave like dormant entities, unable to grow in the presence of the antibiotic but reassuming metabolic functions when the compound disappears. The genetic and biochemical basis of persisters is not completely understood, but it is intuitive that they also represent a foundation for the accumulation of mutations or the acquisition of resistance elements.
The concentration of antibiotic in the medium where bacteria are present is of critical importance in terms of selection of resistance. Mutants of S. pneumoniae or E. coli with different levels of resistance to beta-lactams due to rearrangements in penicillin-binding protein genes or mutations in extended-spectrum beta-lactamase genes, respectively, are selected within a defined range of antibiotic concentrations.42,43 The “mutant selection window” (MSW) varies depending on the antibiotic. The concentration over the MIC at which no mutants are selected is defined as the “mutant prevention concentration” (MPC).44 On the other hand, sub-MICs of (some) antibiotics can also select resistant mutants, by stimulating gene duplication45 and genetic recombination (increasing transfer rates). The importance of low-level resistance is exemplified by qnr genes. These genes code for proteins that cause a moderate increase in the MIC of fluoroquinolones, which although is insufficient to surpass the currently accepted breakpoints for clinical resistance, it favours the selection of mutations in gyrA (alone or combined with mutations in parC), and increases the level of resistance to actual clinical relevance.46
Additionally, some antibiotics (such as quinolones, cotrimoxazole or beta-lactams) can favour the selection of resistant organisms by inducing the SOS response or the direct induction of error-prone DNA polymerases; this increases mutation rates, offering the opportunity for one or more of the new mutations to confer a selective advantage in the presence of the indicated antibiotics.47
Selection can simply work on gene-operons-mobile elements, rather than on a complete bacterium. In these circumstances, the indicated elements become themselves evolutionary units.48
Multiple studies have shown that the emergence of resistant organisms is associated with the use of antibiotics. A good example was the emergence of penicillin resistance in strains of Staphylococcus aureus shortly after the implementation of this antibiotic for clinical use.
In the clinical setting, selecting an organism resistant to a certain antibiotic is not necessarily driven by the considered compound, a consequence of the phenomenon of co-resistance.49 As resistant organisms frequently present resistance to multiple agents, using one agent results in a selective pressure for all other compounds to which the organisms is simultaneously resistant. From another perspective, some antibiotics inhibit normal microbiota, allowing the overgrowth of resistant organisms that, in the absence of the antibiotic, lack an ecological niche in which to develop; this is the case of anti-anaerobes and colonization by glycopeptide-resistant enterococci, or the effect of multiple antibiotics allowing infections by Clostridium difficile.50
Antibiotics can select resistant organisms not only because they are used in human medicine; the existence of a reservoir of mobile resistance genes in food animals was soon recognized in Salmonella enterica serovar. Typhimurium is resistant to tetracyclines, and there are now multiple examples of links between resistant bacteria isolated from animals (e.g., glycopeptide-resistant enterococci, some methicillin-resistant S. aureus, quinolone-resistant or ESBL-producing enterobacteria) and pathogens involved in human colonisation and infection. The Swann Committee to the British Government advised that antibiotics used in human therapy or capable of selecting resistance in human pathogens should not be used as feed additives.51 This was also later adopted in the European Union in relation to antibacterials as growth promotors.
Antibiotic restriction without recovery of bacterial activityThe impact of the use of antimicrobials on resistance to antimicrobials has been tested in some studies.52–54 Gottesman et al. in Israel53 assessed the impact of the restriction of ciprofloxacin use on the resistance profiles of Escherichia coli from urine isolated in the community. After the intervention, a significant reduction (>40%) in quinolone consumption during the studied period was observed, which was associated with a significant decrease (25%) in the isolation of E. coli non-susceptible to quinolones. This effect was reversed when the consumption of quinolones rose again.
Another important example was described by Meyer et al.54 in a nosocomial outbreak at a hospital in New York caused by a strain of Klebsiella pneumoniae resistant to ceftazidime. The authors described an increase of up to 600% in the consumption of ceftazidime in the two years prior to the outbreak, due to the presence of infections caused by Acinetobacter baumannii. After recognition of the outbreak, and the implementation of appropriate measures including a reduction in the consumption of ceftazidime (of up to 80%), concomitant with the use of barrier measures for patients colonized and infected, a clear reduction in the number of cases of K. pneumoniae resistant to ceftazidime was observed. However, the infection control measures that usually tend to be complemented with antimicrobial restriction are an important variable of confusion in this regard.
There are several studies indicating that microorganisms with antimicrobial resistance mechanisms to antibiotics of very limited use in recent decades, sparingly used at present, or even almost in disuse (e.g., chloramphenicol), are still isolated frequently from human infections. An example would be that of Enterobacteriaceae and streptomycin, an antibiotic with very low consumption at present.55 In a study performed in a London hospital, resistance to streptomycin was evaluated in 477 enterobacteria. The authors observed up to 20% resistance to streptomycin in the studied population, and among them, 70% of the antimicrobial resistance was associated with cross-resistance to spectinomycin. The apparent causal explanation of streptomycin resistance was therefore cross-resistance to spectinomycin, whose molecular basis consisted of the gene ant (3”)-Ia included into integron Tn21, where other resistance genes can coexist. Spectinomycin has been and is used especially for the treatment of gonorrhoea, while streptomycin is currently almost restricted to treatment of Mycobacterium tuberculosis infections or in combination with β-lactams for the treatment of enterococci with high resistance to gentamicin. This situation could favour the existence and persistence of resistance to streptomycin despite its limited use in humans.
Along the same lines, Enne et al.,56 in a project of national scope in the United Kingdom, studied the impact of the restriction of sulfonamide use on the prevalence of E. coli resistant to this group of antimicrobials. Between 1991 and 1999, the Royal Hospital in London determined the MIC of sulfamethoxazole (and of a broad panel of antimicrobials) for 350 consecutive clinical isolates of E. coli. Despite the significant decrease in prescriptions of sulfonamides (from 3,208,000 prescriptions per year in 1991 to 77,000 prescriptions per year in 1999), the frequency of antimicrobial resistance remained high, with a prevalence of 39.7% and 46% in 1991 and 1999, respectively. Resistance to sulfonamides is typically associated with the presence of sul type genes, which encode for a dihydropteroate synthase. These genes are regularly found in class I integrons, in plasmids or in the chromosome. When the authors studied the presence of integrons in isolates from the United Kingdom, they found a prevalence of 16.4% (1991) and 17.5% (1999), and they observed an increase in the prevalence of sul genes, from 26.7% to 36.5%, in the same years. In addition, they reported that these genes were encoded on plasmids of large size, some of them conjugative and with a profile of multi-resistance. The conclusion of this study is that despite a large decline in antibiotic prescription, there was no reduction in the resistance. This result may be due to a genetic linkage of the resistance gene to other determinants of resistance in mobile genetic elements, which help maintain resistance to sulfonamides in various environments of selective pressure.
It is also interesting to consider the notion of biological cost or “fitness cost”. This biological concept focuses on the possibility that antimicrobial resistance imposes a cost for replication or transferability of the resistant microorganisms because of the genetic burden needed to maintain the mechanism of resistance. If this is so, once the antibiotic (and therefore selective pressure) disappears, the resistant strain will be replaced by the sensitive one. If the biological cost is minimal or very small, the resistant population may remain without being replaced by the sensitive one. This low biological cost may also represent an explanation (at least in part) for some of the ecological studies detailed above. Similarly, low cost resistance mechanisms explain the presence of resistant bacteria once the corresponding antimicrobial has decreased or disappeared. In relation to routine medical practice, one of the activities referred to in the policy of antimicrobials in hospital-level programs (antimicrobial stewardship), is “antimicrobial cycling”. Basically, this strategy is based on alternating two or more classes of antibiotics for periods of months or years, to reduce or slow the bacterial evolution and spread of resistance to antibiotics in hospitals. Mathematical models have shown, however, that rotation carried out in this way is not completely effective for the proposed objective. However, other approaches, such as “mixing”, where each treated patient receives one of different families of antimicrobials used simultaneously in the hospital, has been predicted to be more effective.57
It must be taken into account that the impact and outcome of an intervention in the use of antibiotics will depend on the location (hospital vs. community), the type of antimicrobial (and the mechanism of resistance to it), and the microorganism itself. According to the discussion throughout the chapter, the reduction or elimination of an antimicrobial will not always translate into an increase in the activity of the same molecule.
Impact of new antibiotic incorporations in bacterial resistanceThe evolution of antimicrobial resistance and a dearth of new antibiotics in the pipeline raise the possibility of untreatable multi-drug resistant (MDR) infections. Recently, there have been some cases of extreme drug-resistant (XDR) bacteria, also known as “superbugs”, which will become more and more common. The WHO recently identified antimicrobial resistance as one of the three greatest threats to human health.58
Why does antibiotic resistance happen? The answer is complex. Levy's theory59 states that the problem of antibiotic-resistance could be bypassed by avoiding the use of antibiotics, a concept challenged by the several studies, as described above.
Resistance was found even to synthetic antibiotics that did not exist on earth until the 20th century. These results underscore a critical reality: antibiotic resistance is already widely disseminated in nature, and to drugs that we have not yet invented.
Antibiotic-resistance has been detected in many bacteria isolated from extreme environments. Widespread resistance to a number of therapeutically useful antibiotics was observed by investigators among bacterial isolates obtained from various types of samples collected from Antarctica, where exposure of the organisms to antibiotics is highly improbable.60 It appears that antibiotics and resistance mechanisms might not have evolved merely as weapons to fight-off antibiotic onslaught, as was believed earlier, but could be integral functions of bacterial physiology.
Over the past 30 years, only two truly novel classes of antibiotics have entered the market: oxazolidinones (linezolid) and cyclic lipopeptides (daptomycin) - and resistance has been already documented for both compounds in the clinical setting.
A recent European Union based study reported in the document ‘The bacterial challenge: time to react’, looked at the additional burden of resistance in terms of attributable mortality and length of hospital stay.60 In Europe, the ECDC reported that 25,000 people die each year from antibiotic-resistant bacteria. The annual cost to the US health care system of antibiotic-resistant infections is already estimated at between US$ 21 billion to US$ 34 billion.61
There is a clear need for new drugs with novel mechanisms of action that can be used to combat the increasing number of bacteria that are resistant to multiple classes of currently available agents. It is obvious that the clinical aspect of antibiotic resistance is only the tip of the iceberg, and most aspects of the study of antibiotic-resistance of bacteria have remained unexplored until now. However, it is the indiscriminate and inappropriate use of antibiotics—in outpatient clinics, amongst hospitalised patients and in the food industry—which is the single largest factor leading to antibiotic resistance. Prudent use of antibiotics might help to slow the emergence of resistant strains, but the strategy cannot ensure complete reversal and disappearance of resistance. Unnecessary and inappropriate use of antibiotics favours the emergence and spread of resistant bacteria. The high prevalence of antimicrobial use (36.2%, ECDC-2012) in primary hospitals could be an important factor in antibiotic resistance.61 Half of all antibiotics consumption may be unnecessary and greatly contributes to increasing bacterial resistance. Paradoxically, underuse through inappropriate choice, inadequate dosing, poor adherence to treatment, and substandard antimicrobials, also plays an important role in the emergence and spread of AMR.
It is helpful that a number of professional and civil organisations, including the WHO, have put forward position papers and recommendations on preserving the beneficial impact of antibiotics.62 The question of how to implement specific actions to protect antibiotics is an important one - a key part of so-called “antibiotic stewardship” - but antimicrobial stewardship by itself cannot alleviate the problem of antimicrobial resistance. If all inappropriate antibiotic use were eliminated, antibiotic-resistant infections would still occur, but at a lower frequency and with clinical impact minimised.
Would it be possible to live in a world without antibiotics? The answer is no, but unfortunately the antibiotic pipeline is running dry. This is happening essentially for two reasons. First, it is difficult to find new antibiotics with novel mechanisms of action. Second, the high cost/benefit and risk/benefit ratio discourages pharmaceutical companies from investment. It is extremely unlikely that incremental changes in these old chemical structures will yield the innovative, safe and effective new antibiotics that society needs.
However, the era when bacterial infections used to be treated with “antibiotics-only” appears to have come to an abrupt end. Basic science research should be expanded to further study antimicrobial resistance mechanisms and epidemiology; to identify new lead compounds; and to develop vaccines, immunotherapies, antibiotic-free plasmids, nanoparticles, nanosized carriers and other technologies to prevent and treat infections in humans and animals.
Today, less than five percent of products in the research and development pipeline are antibiotic drugs. Moreover, the economic imperative for developing new antibiotics should be taken into account. We need to: a) quantify the cost of resistance to healthcare; b) develop cost-benefit models in relation to new antibacterials; and c) introduce economic incentives.
A recent supplementary article of the Infectious Diseases Society of America (IDSA)63 sets out three strategic actions necessary in the development of new antimicrobials: a) refine and define the capability and limitations of PK/PD to predict efficacy and dosing; b) define candidate drugs’ optimal killing parameters and hence dosing; and c) define the ability of various dosing strategies to prevent the emergence of resistance.
A continuous supply of structurally novel antibacterial agents with multiple mode of action is needed to combat the problem of drug resistance in clinical practice.
Innovative incentive schemes are needed to stimulate industry to research and develop new antimicrobial drugs for the future. Incentives need to be acceptable to the pharmaceutical industry, and the role and use of private/public partnerships also needs to be fully explored and adopted. Until a global alliance for antibiotic drug discovery and development is formed, pharmaceutical companies need to recognise that many expensive medicines in their portfolio and in development may be useless if patients succumb to fatal infections. We need to engage with and provide society with the tools to lobby for antibacterials.
Perhaps what is really important is not only how to reduce resistance in the future, but how to manage the resistant organisms already present in our hospitals today. The very complex picture of antibiotic-resistance requires a multi-disciplinary hospital infections advisory group to adequately manage the antimicrobials in hospital settings as proposed in the Spanish scheme “Programa de Optimización de Antibióticos” (PROA).
Conflicts of interestThe authors declare that they have no conflicts of interest.