For the microbiological diagnosis of bacteremia and fungemia, it is essential to use automated blood culture systems that guarantee good performance in the detection of microorganisms.
We evaluated the BT24 system for blood cultures (BCs) by comparing it with the BACTEC™ FX system in detection of positive spiked BCs and bottles, subsequent growth of the microorganism, and time to detection (TTD).
MethodsThe parallel analysis of both systems was performed with 160 strains of 31 different species, each inoculated under the same conditions and simultaneously in six BC bottles.
The Chi-square test was used for comparison per BC and per bottle detection. Wilcoxon test and Student's t-test were used for comparison of TTDs.
ResultsOverall, the following BC (aerobic bottle+anaerobic bottle) were detected as positive: 160/160 (100%) in BACTEC FX and 158/160 (98.7%) in BT24 (p=0.31). Furthermore, the following pediatric BCs (1 single bottle, aerobic) were also detected: 159/160 (99.4%) in BACTEC FX and 156/160 (97.5%) in BT24 (p=0.17).
TTDs were longer with BT24 system than with BACTEC FX system, especially for Staphylococcus aureus strains.
ConclusionsWe consider the BT24 system with good performance and specificity comparable to the reference system, but inferior in TTD.
Para el diagnóstico microbiológico de la bacteriemia y la fungemia es esencial utilizar sistemas automatizados de hemocultivos que garanticen un buen rendimiento en la detección de microorganismos.
Evaluamos el sistema BT24 para hemocultivos comparándolo con el sistema BACTEC™ FX en la detección de hemocultivos y frascos positivos, el crecimiento posterior del microorganismo y el tiempo hasta la detección (TTD).
MétodosEl análisis en paralelo de ambos sistemas se realizó con 160 cepas de 31 especies diferentes, cada una inoculada en las mismas condiciones y simultáneamente en seis botellas de hemocultivos.
Se utilizó la prueba de chi-cuadrado para la comparar la detección por hemocultivo y por botella. La prueba de Wilcoxon y la prueba t de Student se utilizaron para la comparación de los TTD.
ResultadosEn general, se detectaron como positivos los siguientes hemocultivos (frasco aerobio+frasco anaerobio): 160/160 (100%) en BACTEC FX y 158/160 (98,7%) en BT24 (p=0,31). Además, también se detectaron los siguientes hemocultivos pediátricos (un solo frasco, aerobio): 159/160 (99,4%) en BACTEC FX y 156/160 (97,5%) en BT24 (p=0,17).
Los TTD fueron más largos con el sistema BT24 que con el sistema BACTEC FX, especialmente para las cepas de Staphylococcus aureus.
ConclusionesConsideramos que el sistema BT24 tiene buen rendimiento y especificidad, siendo comparable al sistema de referencia pero inferior en TTD.
One of the main causes of mortality in European and North American countries is bloodstream infection (BSI).1 Rapid detection and identification of the causative agent is important to initiate adequate antibiotic therapy and thereby reduce morbidity and mortality.2
Blood cultures (BCs) are the “gold standard” for the microbiological diagnosis of bacteremia and fungemia, so it is essential to use automated systems that guarantee good performance in the detection of microorganisms.3
The BACTEC™ FX system (BACTEC; Becton Dickinson, Sparks, MD, USA) is currently one of the most widely used automated systems in clinical microbiology laboratories worldwide for the detection of microorganisms in blood.3 The positive signal mechanism consists of measuring the amount of CO2 released by growing microorganisms by fluorescence.
Zhuhai DL Biotech (Guandong, China) has developed the BT24 series system which detects microorganisms by measuring the amount of CO2 produced by colorimetry. The BT24 system is easy to operate and similar to the BACTEC FX system. The capacity is variable, the equipment we have used is a module with a capacity of 24 bottles and several modules can be added, which is very useful for small laboratories with a low number of samples. The system consists of a touch screen and availability of connection to the LIS. The bottles are loaded manually as in the BACTEC FX system.
The aerobic and anaerobic bottles of the BT24 system have a volume of 30ml, the blood volume supported is 3–10ml, although it is recommended to add 8–10ml. The pediatric (aerobic) bottles have a volume of 25ml, the sample volume supported is 1–3ml. All three culture media contain resins to neutralize antibiotics. Composition of the culture media and other details can be found at Hardy et al.4
For BSI detection in adult patients, two bottles are used per BC drawn, aerobic bottle for isolation of aerobic and facultative anaerobic bacteria, and anaerobic bottle for isolation of facultative and strict anaerobes. In pediatric patients, the blood collected is inoculated in a single aerobic bottle optimized for small blood volumes.
The aim of this study was to evaluate the BT24 system by analyzing the detection of positive BC bottles and BC, and the subsequent growth of the microorganism, as well as the time to detection (TTD) and comparing it with the BACTEC™ FX system. We considered that the most important parameter for the evaluation is the detection per BC, not per bottle.
Material and methodsThe parallel analysis of the two systems was performed with 160 strains of 31 different species, all of them from BCs samples (Table 1). The number of strains of the different species inoculated in each BC corresponds to a proportion similar to the bacteremias and fungemias that we have in our tertiary hospital over the course of a year.
List of species and number of strains used for the evaluation.
| Specie | Number of strains |
|---|---|
| Gram (−) | |
| Escherichia coli | 16 |
| Klebsiella pneumoniae | 19 |
| Proteus mirabilis | 4 |
| Salmonella enterica | 3 |
| Stenotrophomonas maltophilia | 1 |
| Pseudomonas aeruginosa | 12 |
| Acinetobacter baumanii | 3 |
| Klebsiella aerogenes | 4 |
| Enterobacter cloacae complex | 8 |
| Serratia marcescens | 4 |
| Morganella morganii | 4 |
| Citrobacter freundii | 3 |
| Haemophilus influenzae | 2 |
| Aeromonas sp. | 1 |
| Gram (+) | |
| Staphylococcus aureus | 23 |
| Staphylococcus epidermidis | 8 |
| Staphylococcus intermedius | 1 |
| Streptococcus anginosus | 2 |
| Streptococcus agalactiae | 1 |
| Streptococcus bovis | 1 |
| Streptococcus lugdunensis | 1 |
| Streptococcus pyogenes | 2 |
| Streptococcus pneumoniae | 1 |
| Enterococcus faecalis | 18 |
| Enterococcus faecium | 11 |
| Listeria monocytogenes | 1 |
| Yeast | |
| Candida albicans | 2 |
| Candida tropicalis | 1 |
| Candida glabrata | 1 |
| Candida krusei | 1 |
| Candida parasilopsis | 1 |
Each strain was inoculated under the same conditions and simultaneously in six blood culture bottles (Bactec Plus aerobic/F, Bactec Lytic/10 anaerobic/F and Bactec Peds Plus/F from the BACTEC FX system, and Anaerobic, Aerobic and Children's Blood Culture bottle from the BT24 system). Therefore, we analyzed 160 BC Aerobic/Anaerobic and 160 BC Pediatric, using 480 bottles of BACTEC FX and 480 bottles of BT24.
To simulate real bacteremia conditions, each bottle was inoculated with 5ml, 2.5ml in the pediatric bottles, of human blood and 1ml, 0.5ml in the pediatric bottles, of a solution with 100–200CFU/ml, which was previously prepared by diluting 1/1,000,000 a 0.5 McFarland suspension of each microorganism.5 These bottles were incubated at the same time in both systems.
Subsequently, in the bottles where no growth was detected, a subculture was performed after 5 days of incubation and on the same day of positivity in the bottles where growth was detected.
The Chi-square test was used for comparison per BC and per bottle detection, considering p<0.05 as significant. Wilcoxon test and Student's t-test were used for comparison of TTDs.
A pool of discarded blood is provided to us by our hospital's Blood Bank. The research has been approved by our institute's ethics committee (Comité de Ética para la Investigación con medicamentos, CEIm) prior to conducting the research (CEIm21/39), these experiments were conducted according to established ethical guidelines, and informed consent obtained from the participants was not necessary as we used spiked BCs. The study complies with all regulations.
ResultsOverall, the following BC (aerobic bottle+anaerobic bottle) were detected as positive: 160/160 (100%) in BACTEC FX and 158/160 (98.7%) in BT24 (p=0.31). And the following pediatric cultures (1 single bottle, aerobic): 159/160 (99.4%) in BACTEC FX and 156/160 (97.5%) in BT24 (p=0.17).
Growth was detected in the majority of bottles incubated in both systems, in BATECT FX (475/480, 98.9%) and in BT24 (470/480, 97.9%), with no significant differences in the number of bottles detected as positive (p=0.19). No false negatives were observed, in the negative bottles the microorganism did not grow in the subculture.
By the BACTEC FX system inoculated with Gram-negative species, growth was detected in 250/252 bottles (99.2%). No growth was observed from the subculture of the two anaerobic bottles which were negative. The BT24 system detected growth in 243/252 bottles (96.4%). All the bottles that were negative (3 aerobic, 3 anaerobic and 3 pediatric) contained Escherichia coli strains, and no growth was detected in subsequent subcultures. The BACTEC FX system detected growth in 83/83 (100%) of the aerobic/anaerobic and pediatric BC while the BT24 system detected 82/84 (97.6%) aerobic/anaerobic BC and 81/84 (96.4%) pediatric BC.
In bottles inoculated with Gram-positive species (n=210), growth was detected in all 210 bottles (100%) in both the BACTEC FX system and the BT24 system. Both systems detected 70 (100%) aerobic/anaerobic BC and 70 (100%) pediatric BC.
In the bottles (n=18) inoculated with yeast strains (n=6), 15 were detected as positive by BACTEC FX system (83.3%) while 17 were positive by BT24 system (94.4%). No growth was observed in the subcultures of the bottles in which no growth was detected by both systems. Both systems detected 6 (100%) aerobic/anaerobic BC and 5 (83.3%) pediatric BC.
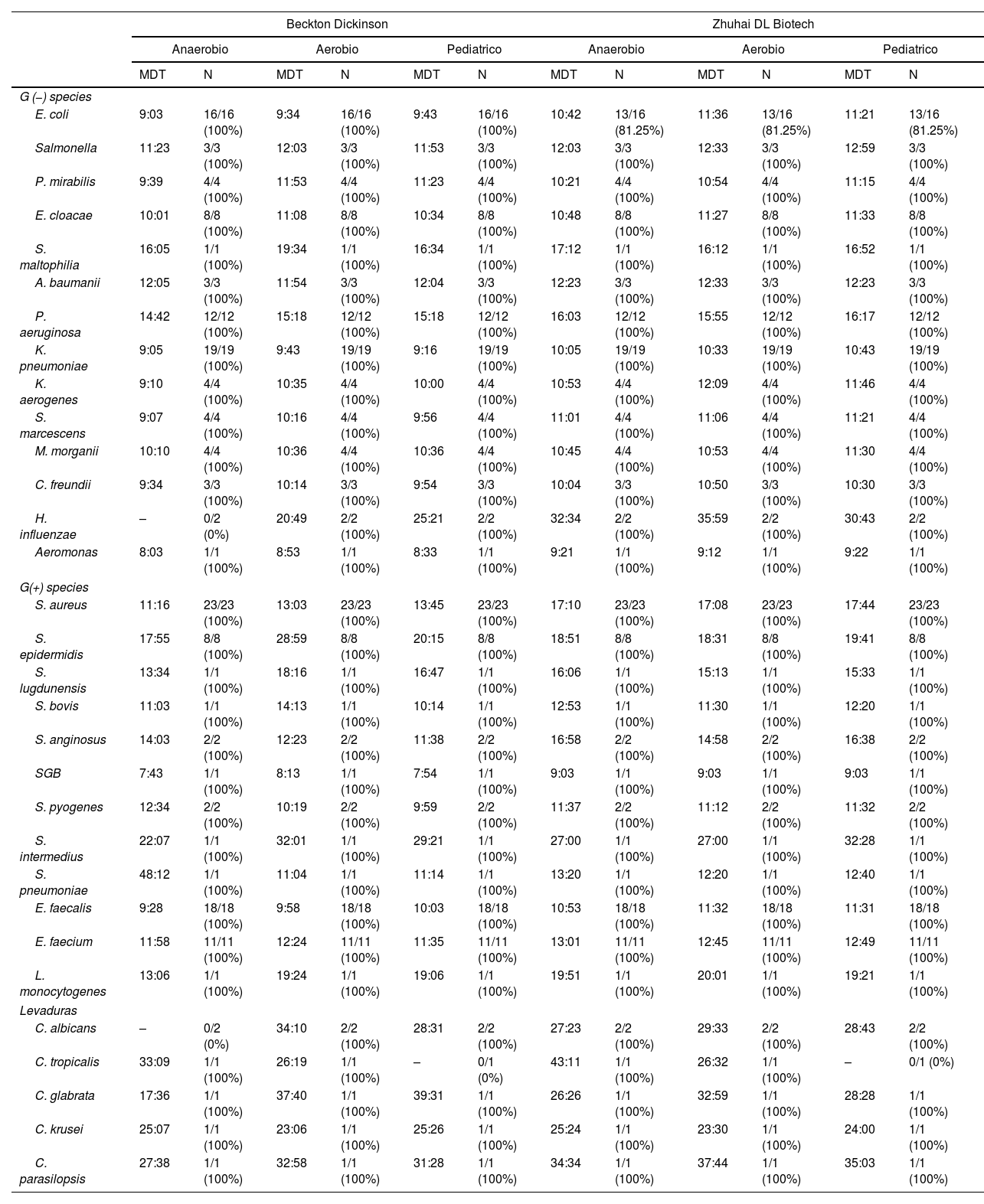
The differences of median TTDs for both systems (Table 2) ranged in general from 0 to 2h, except for Staphylococcus aureus strains, where they were >4h on the BT24 system.
Comparison of time to detection between BACTEC FX system and BT24 system.
| Beckton Dickinson | Zhuhai DL Biotech | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Anaerobio | Aerobio | Pediatrico | Anaerobio | Aerobio | Pediatrico | |||||||
| MDT | N | MDT | N | MDT | N | MDT | N | MDT | N | MDT | N | |
| G (−) species | ||||||||||||
| E. coli | 9:03 | 16/16 (100%) | 9:34 | 16/16 (100%) | 9:43 | 16/16 (100%) | 10:42 | 13/16 (81.25%) | 11:36 | 13/16 (81.25%) | 11:21 | 13/16 (81.25%) |
| Salmonella | 11:23 | 3/3 (100%) | 12:03 | 3/3 (100%) | 11:53 | 3/3 (100%) | 12:03 | 3/3 (100%) | 12:33 | 3/3 (100%) | 12:59 | 3/3 (100%) |
| P. mirabilis | 9:39 | 4/4 (100%) | 11:53 | 4/4 (100%) | 11:23 | 4/4 (100%) | 10:21 | 4/4 (100%) | 10:54 | 4/4 (100%) | 11:15 | 4/4 (100%) |
| E. cloacae | 10:01 | 8/8 (100%) | 11:08 | 8/8 (100%) | 10:34 | 8/8 (100%) | 10:48 | 8/8 (100%) | 11:27 | 8/8 (100%) | 11:33 | 8/8 (100%) |
| S. maltophilia | 16:05 | 1/1 (100%) | 19:34 | 1/1 (100%) | 16:34 | 1/1 (100%) | 17:12 | 1/1 (100%) | 16:12 | 1/1 (100%) | 16:52 | 1/1 (100%) |
| A. baumanii | 12:05 | 3/3 (100%) | 11:54 | 3/3 (100%) | 12:04 | 3/3 (100%) | 12:23 | 3/3 (100%) | 12:33 | 3/3 (100%) | 12:23 | 3/3 (100%) |
| P. aeruginosa | 14:42 | 12/12 (100%) | 15:18 | 12/12 (100%) | 15:18 | 12/12 (100%) | 16:03 | 12/12 (100%) | 15:55 | 12/12 (100%) | 16:17 | 12/12 (100%) |
| K. pneumoniae | 9:05 | 19/19 (100%) | 9:43 | 19/19 (100%) | 9:16 | 19/19 (100%) | 10:05 | 19/19 (100%) | 10:33 | 19/19 (100%) | 10:43 | 19/19 (100%) |
| K. aerogenes | 9:10 | 4/4 (100%) | 10:35 | 4/4 (100%) | 10:00 | 4/4 (100%) | 10:53 | 4/4 (100%) | 12:09 | 4/4 (100%) | 11:46 | 4/4 (100%) |
| S. marcescens | 9:07 | 4/4 (100%) | 10:16 | 4/4 (100%) | 9:56 | 4/4 (100%) | 11:01 | 4/4 (100%) | 11:06 | 4/4 (100%) | 11:21 | 4/4 (100%) |
| M. morganii | 10:10 | 4/4 (100%) | 10:36 | 4/4 (100%) | 10:36 | 4/4 (100%) | 10:45 | 4/4 (100%) | 10:53 | 4/4 (100%) | 11:30 | 4/4 (100%) |
| C. freundii | 9:34 | 3/3 (100%) | 10:14 | 3/3 (100%) | 9:54 | 3/3 (100%) | 10:04 | 3/3 (100%) | 10:50 | 3/3 (100%) | 10:30 | 3/3 (100%) |
| H. influenzae | – | 0/2 (0%) | 20:49 | 2/2 (100%) | 25:21 | 2/2 (100%) | 32:34 | 2/2 (100%) | 35:59 | 2/2 (100%) | 30:43 | 2/2 (100%) |
| Aeromonas | 8:03 | 1/1 (100%) | 8:53 | 1/1 (100%) | 8:33 | 1/1 (100%) | 9:21 | 1/1 (100%) | 9:12 | 1/1 (100%) | 9:22 | 1/1 (100%) |
| G(+) species | ||||||||||||
| S. aureus | 11:16 | 23/23 (100%) | 13:03 | 23/23 (100%) | 13:45 | 23/23 (100%) | 17:10 | 23/23 (100%) | 17:08 | 23/23 (100%) | 17:44 | 23/23 (100%) |
| S. epidermidis | 17:55 | 8/8 (100%) | 28:59 | 8/8 (100%) | 20:15 | 8/8 (100%) | 18:51 | 8/8 (100%) | 18:31 | 8/8 (100%) | 19:41 | 8/8 (100%) |
| S. lugdunensis | 13:34 | 1/1 (100%) | 18:16 | 1/1 (100%) | 16:47 | 1/1 (100%) | 16:06 | 1/1 (100%) | 15:13 | 1/1 (100%) | 15:33 | 1/1 (100%) |
| S. bovis | 11:03 | 1/1 (100%) | 14:13 | 1/1 (100%) | 10:14 | 1/1 (100%) | 12:53 | 1/1 (100%) | 11:30 | 1/1 (100%) | 12:20 | 1/1 (100%) |
| S. anginosus | 14:03 | 2/2 (100%) | 12:23 | 2/2 (100%) | 11:38 | 2/2 (100%) | 16:58 | 2/2 (100%) | 14:58 | 2/2 (100%) | 16:38 | 2/2 (100%) |
| SGB | 7:43 | 1/1 (100%) | 8:13 | 1/1 (100%) | 7:54 | 1/1 (100%) | 9:03 | 1/1 (100%) | 9:03 | 1/1 (100%) | 9:03 | 1/1 (100%) |
| S. pyogenes | 12:34 | 2/2 (100%) | 10:19 | 2/2 (100%) | 9:59 | 2/2 (100%) | 11:37 | 2/2 (100%) | 11:12 | 2/2 (100%) | 11:32 | 2/2 (100%) |
| S. intermedius | 22:07 | 1/1 (100%) | 32:01 | 1/1 (100%) | 29:21 | 1/1 (100%) | 27:00 | 1/1 (100%) | 27:00 | 1/1 (100%) | 32:28 | 1/1 (100%) |
| S. pneumoniae | 48:12 | 1/1 (100%) | 11:04 | 1/1 (100%) | 11:14 | 1/1 (100%) | 13:20 | 1/1 (100%) | 12:20 | 1/1 (100%) | 12:40 | 1/1 (100%) |
| E. faecalis | 9:28 | 18/18 (100%) | 9:58 | 18/18 (100%) | 10:03 | 18/18 (100%) | 10:53 | 18/18 (100%) | 11:32 | 18/18 (100%) | 11:31 | 18/18 (100%) |
| E. faecium | 11:58 | 11/11 (100%) | 12:24 | 11/11 (100%) | 11:35 | 11/11 (100%) | 13:01 | 11/11 (100%) | 12:45 | 11/11 (100%) | 12:49 | 11/11 (100%) |
| L. monocytogenes | 13:06 | 1/1 (100%) | 19:24 | 1/1 (100%) | 19:06 | 1/1 (100%) | 19:51 | 1/1 (100%) | 20:01 | 1/1 (100%) | 19:21 | 1/1 (100%) |
| Levaduras | ||||||||||||
| C. albicans | – | 0/2 (0%) | 34:10 | 2/2 (100%) | 28:31 | 2/2 (100%) | 27:23 | 2/2 (100%) | 29:33 | 2/2 (100%) | 28:43 | 2/2 (100%) |
| C. tropicalis | 33:09 | 1/1 (100%) | 26:19 | 1/1 (100%) | – | 0/1 (0%) | 43:11 | 1/1 (100%) | 26:32 | 1/1 (100%) | – | 0/1 (0%) |
| C. glabrata | 17:36 | 1/1 (100%) | 37:40 | 1/1 (100%) | 39:31 | 1/1 (100%) | 26:26 | 1/1 (100%) | 32:59 | 1/1 (100%) | 28:28 | 1/1 (100%) |
| C. krusei | 25:07 | 1/1 (100%) | 23:06 | 1/1 (100%) | 25:26 | 1/1 (100%) | 25:24 | 1/1 (100%) | 23:30 | 1/1 (100%) | 24:00 | 1/1 (100%) |
| C. parasilopsis | 27:38 | 1/1 (100%) | 32:58 | 1/1 (100%) | 31:28 | 1/1 (100%) | 34:34 | 1/1 (100%) | 37:44 | 1/1 (100%) | 35:03 | 1/1 (100%) |
MDT: median detection times; N: positive bottles/total bottles.
In the species with more than 10 strains tested the differences of median TTD for both systems and the statistical significance are in Table 3.
Results of Wilcoxon test and Student's t-test.
| Species | No. strains | Statistical differences |
|---|---|---|
| K. pneumoniae | 19 | p<0.002 |
| P. aeruginosa | 12 | p<0.016 |
| E. coli | 13 | p<0.002 |
| E. cloacae | 8 | p<0.018 |
| E. faecalis | 18 | p<0.003 |
| E. faecium | 11 | p<0.002 |
| S. aureus | 23 | p<0.001 |
All results in favor of the BACTEC FX system.
BACTEC FX is one of the most used blood culture systems over the world.
The BT24 and BACTEC FX automated systems were compared by simulating BC of adult or pediatric patients, inoculating different bacterial and yeast strains in anaerobic/aerobic and pediatric bottles of both systems, incubating them in parallel and analyzing the growth and the TTD of each bottle.
We used healthy human blood to better simulate BCs routinely collected from patients. Our simulation also relied on two different volumes of blood in aerobic/anaerobic or pediatric BC bottles to, respectively, model adult and pediatric cultures.
The global detection rate did not differ significantly in the number of positive BC in BATECT FX and BT24.
Significant differences on TTD were observed for several species, in particular for S. aureus strains, which are of high relevance in bacteremias.
In a comparison of BACTEC FX with BACT/ALERT VIRTUO (bioMérieux), this last system allowed faster growth detection for most of the 33 species tested.3
In a recent comparison of several blood culture system from China using as a reference the BacT/ALERT 3D (bioMérieux), DL-biotech DL 60 had satisfactory performance.4 The software of DL-biotech DL60 has been incorporated to BT24, probably improving its performance.
Delays in appropriate antimicrobial treatment were associated with increased 30-day mortality after 12h from blood culture collection, but not at 1, 3, and 6h, in BSI.6 BT24 could be useful in countries with limited resources, as it is an affordable system.
A limitation of this study is the use of BC simulations, so it cannot be ruled out that performance differs in the presence of antibiotics and other inhibitory factors in clinical samples. The effect of polymicrobial growth was also not evaluated.
Strengths of our study are the considerable number of strains of various species representative of the causative agents of bacteremia in our hospital tested, and the use of human blood.
As conclusions, the majority of aerobic/anaerobic and pediatric BCs were detected with the BT24 system. TTDs were longer with BT24 than with BACTEC FX system, especially for S. aureus strains. Therefore, we consider the BT24 with good performance and specificity comparable to the reference system, but inferior in TTD.
Controlled studies are warranted to evaluate the BT24 system in clinical samples.
FundingThis work was supported by Zhuhai DL Biotech. In addition to providing reagents and funding for this study, Zhuhai DL Biotech did not participate in the design of the study.
Authors’ contributionsAll authors contributed to the study conception and design. All authors read and approved the final manuscript.
Competing interestsThe authors have no relevant financial interests to disclose.
A special thanks to Israel J. Thuissard and Cristina Andreu for helping with statistical calculations.