EUCAST breakpoints are more restrictive than those defined by CLSI. This study highlights the discrepancies between CLSI and EUCAST in a well characterized isogenic Escherichia coli collection and their correlations with specific quinolone resistance mechanisms. The greatest number of discrepancies was observed in strains containing 2–4 resistance mechanisms (MIC values on the borderline of clinical resistance). Bearing in mind that quinolones are concentration dependent antimicrobial agents, small changes in MIC may have relevant consequences for treatment outcomes.
Los puntos de corte de EUCAST son más restrictivos que los definidos por CLSI. Este estudio analiza las discrepancias entre CLSI y EUCAST en una colección isogénica de Escherichia coli y su correlación con mecanismos específicos de resistencia a quinolonas. El mayor número de discrepancias se observó en cepas que contienen 2–4 mecanismos de resistencia (con CMI en el límite de la resistencia clínica). Teniendo en cuenta que las quinolonas son agentes antimicrobianos concentración-dependientes, pequeños cambios en el valor de la CMI pueden tener consecuencias relevantes para el resultado del tratamiento.
Fluoroquinolones are broad-spectrum antimicrobial agents frequently used in clinical practice. Fluoroquinolone resistance rates have greatly increased over recent decades. In Enterobacteriaceae, quinolone resistance is primarily related to mutations in chromosomal genes encoding the quinolone targets DNA gyrase and topoisomerase IV, and secondly due to loss of porins or overexpression of efflux pumps like AcrAB-TolC (which is negatively regulated by marR).1 Three known mechanisms have been reported so far in the last 15 years in connection with the emergence of plasmid-mediated quinolone resistance (PMQR) mechanisms: qnr genes encoding pentapeptide repeat proteins; aac(6′)-Ib-cr encoding an aminoglycoside acetyltransferase variant; and qepA or oqxAB, encoding active efflux pumps.2
The Clinical and Laboratory Standards Institute (CLSI) and the European Committee on Antimicrobial Susceptibility Testing (EUCAST) use different criteria for defining clinical breakpoints for fluoroquinolones in Enterobacteriaceae,3,4 with EUCAST breakpoints being more restrictive than those defined by CLSI. PMQR genes themselves confer low-level quinolone resistance, and their effect on the selection of quinolone resistance in association with other mechanisms has been reported.5,6 A combination of qnrA1, qnrB1, qnrC, qnrD1 and qnrS1 genes together with a Ser83Leu substitution and/or Asp87Asn substitution in GyrA and/or Ser80Arg substitution in ParC and/or marR gene deletion using an Escherichia coli ATCC 25922 model was recently analyzed.6 In that study, at least four resistance mechanisms [combination of Ser83Leu substitution, Ser80Arg substitution, other chromosomal modifications (Asp87Asn substitution or marR deletion) and qnr expression] were necessary to produce a clinically resistant phenotype according to CLSI breakpoints (MIC of ciprofloxacin >2mg/L). In the present study, we have extended our previous results6,7 and highlight the discrepancies between CLSI and EUCAST observed in this isogenic E. coli collection, and their correlations with specific molecular mechanisms.
MethodsSixty isogenic E. coli strains (based on E. coli ATCC 25922 strain), harboring the most frequently found chromosomal mutations associated with FQ resistance in the gyrA and/or parC genes and/or a deletion in the marR gene were combined with qnr genes for susceptibility testing to quinolones.6
Minimal inhibitory concentrations (MIC) were previously determined in triplicate for each bacterial strain using the broth microdilution method, according to CLSI reference methods.6 Quinolones analyzed were: ciprofloxacin (CIP, Sigma–Aldrich), levofloxacin (LVX, Sigma–Aldrich), norfloxacin (NFX, Sigma–Aldrich) and ofloxacin (OFX, Sigma–Aldrich). Stock solutions were prepared in accordance with Clinical and Laboratory Standards Institute (CLSI) guidelines (www.clsi.org).3
Results and discussionThe therapeutic relevance of the acquisition of qnr genes to the bactericidal activity of fluoroquinolones remains unclear. Qnr proteins provide low-level quinolone resistance facilitating the in vitro emergence of higher levels of resistance in the presence of quinolones at therapeutic levels.6,8 Previous in vivo studies have shown that the presence of qnr genes in association with chromosomal mechanisms of quinolone resistance seems to be significant for the in vivo activity of fluoroquinolones,9 indicating that small changes in MIC, like those conferred by qnr genes, could be significant.
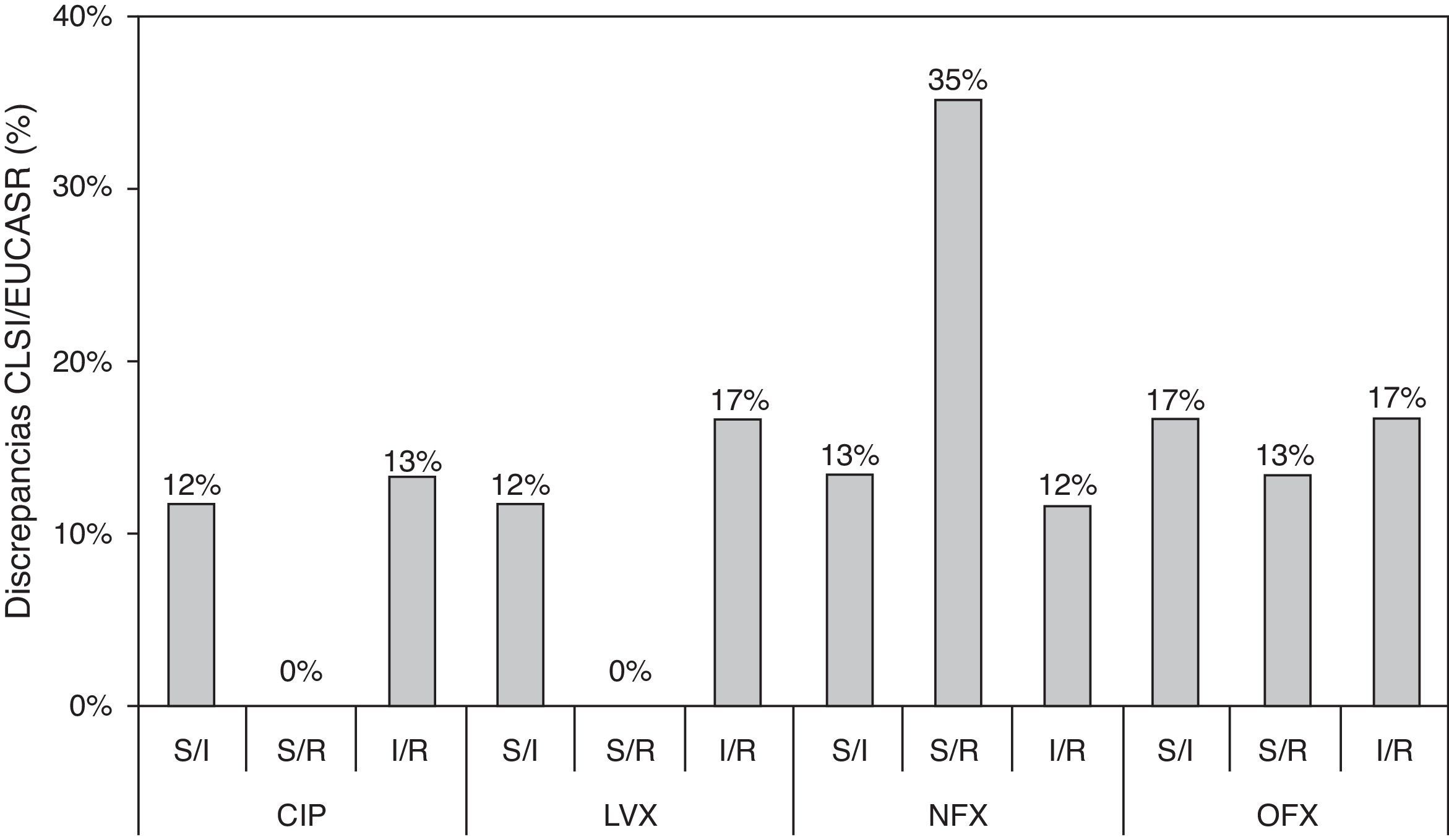
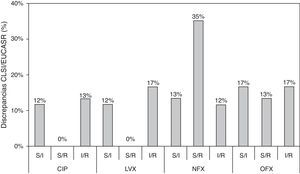
In the present study, we show an additional analysis of an isogenic collection,6 highlighting clear discrepancies between the CLSI and EUCAST clinical categories and establishing a correlation with the molecular mechanisms implicated. Three types of discrepancy were observed when CLSI and EUCAST criteria were compared: 32 susceptible/intermediate susceptibility (S/I); 29 susceptible/resistance (S/R); and 35 intermediate susceptibility/resistance (I/R) (Table S1). MIC values that were close to category breakpoints were responsible for the observed discrepancies; the corresponding phenotypes have been associated with a higher frequency of selection of quinolone-resistant mutants in strains harboring PMQR genes and/or chromosomal mechanisms.6,8 Discrepancies between CLSI and EUCAST breakpoints were always found in strains containing at least two mechanisms in combination and when qnr genes were the only resistance mechanism (except for qnrD1). Discrepancies according to fluoroquinolone tested were observed in 25%, 29%, 60% and 47% of the whole collection for ciprofloxacin, levofloxacin, norfloxacin and ofloxacin, respectively (Fig. 1). A greater number of discrepancies were observed for norfloxacin and ofloxacin, with a 35% S/R discrepancy rate for norfloxacin. The greatest number of discrepancies was observed in strains containing 2–4 resistance mechanisms, also dependent on the fluoroquinolone tested. The presence of qnr genes, in the absence of chromosomal mechanisms of quinolone resistance, only originated discrepancies in norfloxacin and ofloxacin clinical categories. Morgan-Linnell et al. reported that the combined effect of DNA gyrase and topoisomerase IV mutations on fluoroquinolone resistance in isogenic E. coli C600 strains showed that at least three mutations—two of which had to be in gyrA—were necessary to exceed CLSI resistance breakpoints.10 Strains with two substitutions, one in gyrA and one in parC, were susceptible to fluoroquinolones, with ciprofloxacin MICs of <1mg/L. On the other hand, in a model derived from E. coli MG1655, Marcusson et al. showed that strains with two mutations in type II topoisomerase genes presented intermediated susceptibility to ciprofloxacin according to CLSI breakpoints.11 Discrepancies between different studies could be partially explained by the MICs of ciprofloxacin in the wild type strains used, with E. coli MG1655 (derived from K12) being higher than E. coli ATCC 25922 (0.016mg/L vs 0.004mg/L, respectively), and support that unknown low level resistance mechanisms could be present in E. coli.12
Discrepancies between clinical categories according to CLSI and EUCAST breakpoints. S, susceptible; I, intermediate susceptible; R, resistant. Comparison of MIC values and CLSI and EUCAST clinical categories, respectively. Clinical breakpoints (mg/L) of fluoroquinolones for Enterobacteriaceae as proposed by CLSI and EUCAST are as follows: CLSI: CIP, susceptible≤1mg/L, intermediate=2mg/L and resistant≥4mg/L; LVX, susceptible≤2mg/L, intermediate=4mg/L and resistant≥8mg/L; NFX, susceptible≤4mg/L, intermediate=8mg/L and resistant≥16mg/L; and OFX, susceptible≤2mg/L, intermediate=4mg/L and resistant≥8mg/L. EUCAST: CIP, susceptible≤0.5mg/L, intermediate=1mg/L and resistant≥2mg/L; LVX, susceptible≤1mg/L, intermediate=2mg/L and resistant≥4mg/L; NFX, susceptible≤0.5mg/L, intermediate=1mg/L and resistant≥2mg/L; and OFX, susceptible≤0.5mg/L, intermediate=1mg/L and resistant≥2mg/L.
Finally, in this study using a set of sixty isogenic E. coli strains,6 significant differences in terms of clinical category were identified when CLSI and EUCAST breakpoints were compared. These differences were particularly relevant in isolates with MIC values on the borderline of resistance according to CLSI or EUCAST breakpoints. These low level resistant phenotypes (breakpoint borderlines) could have clinical relevance leading to in vivo resistance.13 Bearing in mind that the pharmacokinetic parameter for the clinical efficacy of quinolones is the AUC/MIC, small changes in MIC may be significant for treatment outcome.
FundingThis work was supported by the Ministerio de Sanidad y Consumo, Instituto de Salud Carlos III (project PI11-00934) and the Consejería de Innovación Ciencia y Empresa, Junta de Andalucía (P11-CTS-7730), Spain, the Plan Nacional de I+D+i 2008–2011 and Instituto de Salud Carlos, Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Economía y Competitividad, Spanish Network for Research in Infectious Diseases (REIPI RD12/0015) – co-financed by European Development Regional Fund “A way to achieve Europe” ERDF.
Conflict of interestThe authors declare no conflicts of interest.