We characterized AmpC β-lactamase mutations that resulted in ceftolozane/tazobactam resistance in extensively drug-resistant (XDR) Pseudomonas aeruginosa isolates recovered from patients treated with this agent from June 2016 to December 2018.
MethodsFive pairs of ceftolozane/tazobactam susceptible/resistant P. aeruginosa XDR isolates were included among a total of 49 patients treated. Clonal relationship among isolates was first evaluated by pulsed-field gel electrophoresis (PFGE). Multilocus sequence typing (MLST) was further performed. AmpC mutations were investigated by PCR amplification of the blaPDC gene followed by sequencing.
ResultsThe ST175 high-risk clone was detected in four of the pairs of isolates and the ST1182 in the remaining one. All resistant isolates showed a mutation in AmpC: T96I in two of the isolates, and E247K, G183V, and a deletion of 19 amino acids (G229–E247) in the other three. The G183V mutation had not been described before. The five isolates resistant to ceftolozane/tazobactam showed cross-resistance to ceftazidime/avibactam and lower MICs of imipenem and piperacillin/tazobactam than the susceptible isolates.
ConclusionsCeftolozane/tazobactam resistance was associated in all of the cases with AmpC mutations, including a novel mutation (G183V) not previously described. There is a vital need for surveillance and characterization of emerging ceftolozane/tazobactam resistance, in order to preserve this valuable antipseudomonal agent.
Se han caracterizado las mutaciones en la betalactamasa AmpC que han producido resistencia a ceftolozano/tazobactam en aislados de Pseudomonas aeruginosa extremadamente resistente (XDR) en pacientes tratados con este agente desde junio de 2016 hasta diciembre de 2018.
MétodosSe incluyeron 5 pares de aislados (sensibles/resistentes a ceftolozano/tazobactam) de P. aeruginosa XDR entre un total de 49 pacientes tratados. Se estudió la relación clonal mediante electroforesis en campo pulsado y MLST. Las mutaciones en AmpC se caracterizaron mediante amplificación por PCR del gen blaPDC y posterior secuenciación.
ResultadosSe detectó el clon de alto riesgo ST175 en 4 pares de aislados y el ST1182 en el restante. Todos los aislados resistentes mostraron una mutación en AmpC: T96I en 2 aislados, E247K, G183V y una deleción de 19 aminoácidos (G229-E247) en los otros 3. La mutación G183V no había sido descrita antes. Los 5 aislados resistentes a ceftolozano/tazobactam mostraron resistencia cruzada a ceftazidima/avibactam y CMI inferiores de imipenem y piperacilina/tazobactam que los aislados sensibles.
ConclusionesLa resistencia a ceftolozano/tazobactam se asoció con mutaciones en AmpC en todos los casos, incluida una nueva mutación G183V no descrita con anterioridad. La vigilancia y caracterización de la resistencia emergente a ceftolozano/tazobactam es de gran importancia para preservar este nuevo agente antipseudomónico.
Infections due to multridug-resistant (MDR) or extensively drug-resistant (XDR) Pseudomonas aeruginosa strains are associated with nosocomial infections and with significant morbidity and mortality.1 Ceftolozane/tazobactam is a combination of a novel cephalosporin with the β-lactamase inhibitor tazobactam, approved for the treatment of complicated intra-abdominal infections, complicated urinary tract infections, and hospital-acquired/ventilator-associated bacterial pneumonia.2,3 Ceftolozane/tazobactam constitutes a valuable treatment option for MDR and XDR P. aeruginosa infections.2,4 Unlike the previous-generation cephalosporins, ceftolozane/tazobactam has demonstrated increased stability to AmpC β-lactamases2 but since the introduction of this agent in the clinical setting, emerging resistance during therapy has been observed, mainly associated with AmpC mutations.5–7
The aim of this study was to characterize the AmpC β-lactamase mutations leading to ceftolozane/tazobactam emerging resistance in P. aeruginosa XDR isolates recovered from patients treated with this agent.
Material and methodsClinical strainsFive pairs of ceftolozane/tazobactam susceptible/resistant P. aeruginosa XDR isolates were included, obtained from five patients treated with ceftolozane/tazobactam from June 2016 to December 2018 at our hospital. During this period, 49 patients with P. aeruginosa infections (48 P. aeruginosa XDR) were treated with ceftolozane/tazobactam. They presented mainly with respiratory infections (n=18), urinary infections (n=13), and bacteremia (n=9).
Clinical samples were cultured and incubated overnight at 37°C and bacterial identification was performed by MALDI-TOF MS (Bruker Daltonics).
Susceptibility testingAntimicrobial susceptibility of piperacillin/tazobactam, ceftazidime, cefepime, aztreonam, ceftolozane/tazobactam, ceftazidime/avibactam, imipenem, meropenem, tobramycin, amikacin, ciprofloxacin and colistin was determined by broth microdilution (Sensititre®). EUCAST criteria (v6.0 in 2016, v7.1 in 2017, and v8.0 and v8.1 in 2018; http://www.eucast.org) were applied. MDR profile was defined as acquired non-susceptibility to at least one agent in three or more antimicrobial categories, and XDR profile was defined as non-susceptibility to at least one agent in all but two or fewer antimicrobial categories according to Magiorakos et al.8 In all the cases the ceftolozane/tazobactam susceptible isolate was analyzed before the start of the treatment and the ceftolozane/tazobactam resistant isolate during or after the treatment.
AmpC overproduction/detection of carbapenemasesAmpC overproduction was confirmed by the cloxacillin inhibition test with ceftazidime disks. The presence of carbapenemases in the ceftolozane/tazobactam resistant isolates was ruled out through the Neo-Rapid CARB Kit (Rosco Diagnostica). In addition, the presence of OXA-2 and/or OXA-10 β-lactamases was analyzed by PCR as previously described.5,9
Molecular epidemiologyClonal relatedness among isolates was first evaluated by Spe1 pulsed-field gel electrophoresis (PFGE) according to standard protocols.10 Representative isolates from each unique macrorestriction pattern were further analyzed by multilocus sequence typing (MLST) following previously described protocols.11 Isolates were assigned to a sequence type (ST) number according to the allelic profiles available in the MLST Database (http://pubmlst.org/paeruginosa). AmpC mutations were investigated by phenotypic and molecular methods by PCR amplification of blaPDC followed by sequencing with the specific primers AmpC F (5′-ACGACAAAGGACGCCAATCC-3′) and AmpC R (5′-TCAGCGCTTCAGCGGCACC-3′).12
Genbank accession numbersThe nucleotide sequence from isolate 4b described in results (Table 1) has been deposited in the GenBank database under accession number MN525567 (blaPDC-388).
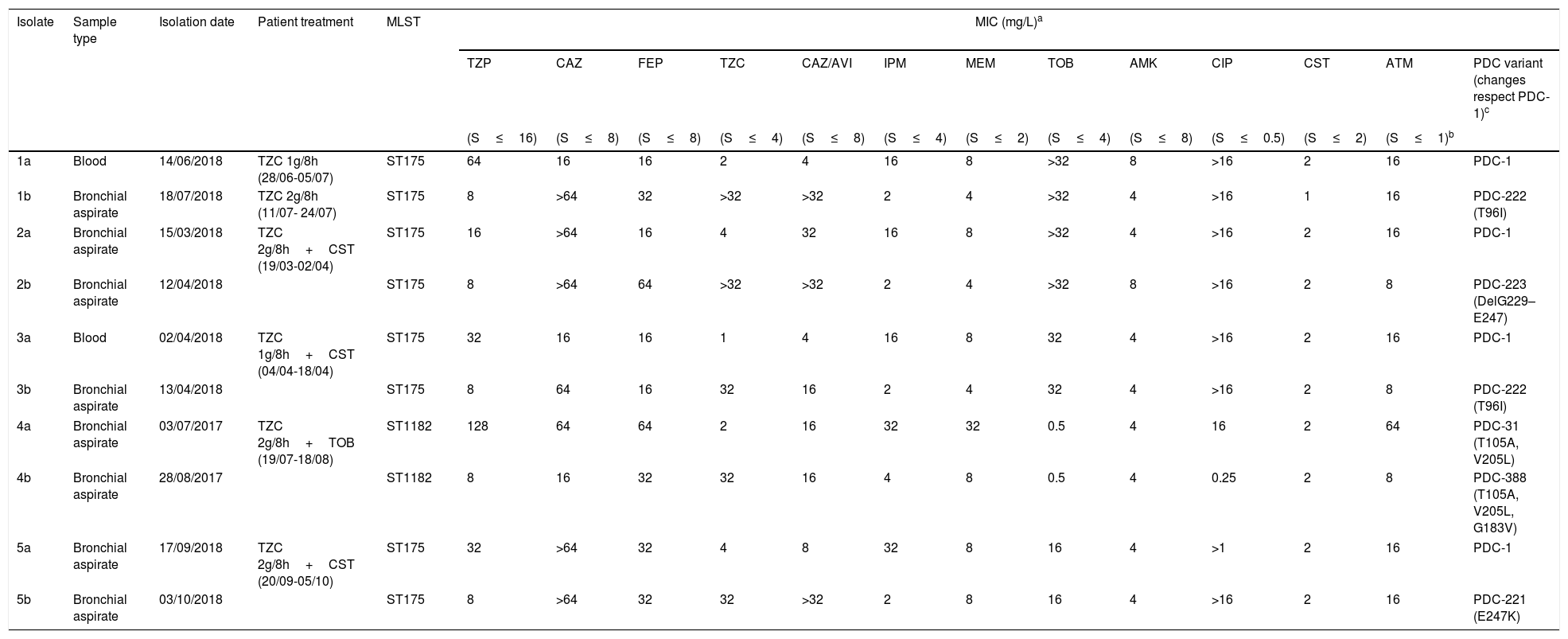
Antimicrobial susceptibility, molecular characterization and AmpC mutations in the five pairs of ceftolozane/tazobactam susceptible/resistant P. aeruginosa isolates.
| Isolate | Sample type | Isolation date | Patient treatment | MLST | MIC (mg/L)a | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TZP | CAZ | FEP | TZC | CAZ/AVI | IPM | MEM | TOB | AMK | CIP | CST | ATM | PDC variant (changes respect PDC-1)c | |||||
| (S≤16) | (S≤8) | (S≤8) | (S≤4) | (S≤8) | (S≤4) | (S≤2) | (S≤4) | (S≤8) | (S≤0.5) | (S≤2) | (S≤1)b | ||||||
| 1a | Blood | 14/06/2018 | TZC 1g/8h (28/06-05/07) | ST175 | 64 | 16 | 16 | 2 | 4 | 16 | 8 | >32 | 8 | >16 | 2 | 16 | PDC-1 |
| 1b | Bronchial aspirate | 18/07/2018 | TZC 2g/8h (11/07- 24/07) | ST175 | 8 | >64 | 32 | >32 | >32 | 2 | 4 | >32 | 4 | >16 | 1 | 16 | PDC-222 (T96I) |
| 2a | Bronchial aspirate | 15/03/2018 | TZC 2g/8h+CST (19/03-02/04) | ST175 | 16 | >64 | 16 | 4 | 32 | 16 | 8 | >32 | 4 | >16 | 2 | 16 | PDC-1 |
| 2b | Bronchial aspirate | 12/04/2018 | ST175 | 8 | >64 | 64 | >32 | >32 | 2 | 4 | >32 | 8 | >16 | 2 | 8 | PDC-223 (DelG229–E247) | |
| 3a | Blood | 02/04/2018 | TZC 1g/8h+CST (04/04-18/04) | ST175 | 32 | 16 | 16 | 1 | 4 | 16 | 8 | 32 | 4 | >16 | 2 | 16 | PDC-1 |
| 3b | Bronchial aspirate | 13/04/2018 | ST175 | 8 | 64 | 16 | 32 | 16 | 2 | 4 | 32 | 4 | >16 | 2 | 8 | PDC-222 (T96I) | |
| 4a | Bronchial aspirate | 03/07/2017 | TZC 2g/8h+TOB (19/07-18/08) | ST1182 | 128 | 64 | 64 | 2 | 16 | 32 | 32 | 0.5 | 4 | 16 | 2 | 64 | PDC-31 (T105A, V205L) |
| 4b | Bronchial aspirate | 28/08/2017 | ST1182 | 8 | 16 | 32 | 32 | 16 | 4 | 8 | 0.5 | 4 | 0.25 | 2 | 8 | PDC-388 (T105A, V205L, G183V) | |
| 5a | Bronchial aspirate | 17/09/2018 | TZC 2g/8h+CST (20/09-05/10) | ST175 | 32 | >64 | 32 | 4 | 8 | 32 | 8 | 16 | 4 | >1 | 2 | 16 | PDC-1 |
| 5b | Bronchial aspirate | 03/10/2018 | ST175 | 8 | >64 | 32 | 32 | >32 | 2 | 8 | 16 | 4 | >16 | 2 | 16 | PDC-221 (E247K) | |
Table 1 shows the antimicrobial susceptibility profiles and the molecular characterization of the ceftolozane/tazobactam susceptible/resistant P. aeruginosa isolates studied. Ceftolozane/tazobactam resistant isolates were found to be identical to their susceptible counterparts by PFGE. Two PFGE macrorestriction patterns were obtained, one in four pairs of isolates, which were further assigned to the ST175 high-risk clone by MLST, and the other in the remaining pair of isolates, assigned to the ST1182.
The five ceftolozane-resistant isolates presented a negative result on the Neo-Rapid CARB kit, excluding the presence of carbapenemases, and a positive cloxacillin inhibition test, indicating the AmpC overproduction. Likewise, none of the isolates were positive for OXA-2 or OXA-10 β-lactamases.
In the four cases belonging to ST175 the initial isolate was only susceptible to amikacin, colistin and ceftolozane/tazobactam, and also to ceftazidime/avibactam in three cases. In the case of ST1182, the initial isolate was susceptible to amikacin, tobramicin, colistin and ceftolozane/tazobactam. Ceftolozane/tazobactam MICs increased from 1 to 4mg/L in the susceptible isolates to ≥32mg/L in the subsequent resistant isolates. Additionally, the five isolates resistant to ceftolozane/tazobactam showed cross-resistance to ceftazidime/avibactam and all showed lower MICs of imipenem, meropenem and piperacillin/tazobactam than the susceptible isolates (Table 1).
All resistant isolates showed a mutation in the AmpC gene not present in the susceptible isolates: T96I in two isolates, and G183V, E247K and a deletion of 19 amino acids (G229–E247) respectively in the other three isolates (Table 1). The G183V mutation is a novel mutation not described before (PDC-388, deposited to GeneBank, accession number MN525567).
The clinical aspects of this cohort are not the object of this study. Briefly, two patients presented bacteremia treated with standard doses of ceftolozane/tazobactam (1g/8h) and three patients presented ventilator associated pneumonia, treated with high doses of ceftolozane/tazobactam (2g/8h). Subsequent ceftolozane/tazobactam resistant isolates were all isolated from bronchial aspirates (Table 1).
DiscussionCeftolozane/tazobactam is a novel cephalosporin with potent in vitro activity against P. aeruginosa and is stable against the most common resistance mechanisms as the overexpression of the chromosomal cephalosporinase AmpC or efflux pumps.2,12,13 In addition, it has been showed to be an effective and safe drug for treating different types of P. aeruginosa infections including these caused by MDR and XDR isolates and those with initially off-label indications as blood stream infections and pneumonia.14 Although ceftolozane/tazobactam has been successfully used as the treatment for these infections, several studies have documented resistance to ceftolozane/tazobactam associated with mutations in the AmpC gene.5,6,14,15 The percentage of ceftolozane/tazobactam resistance development ranged from 2.9% (3 patients out of 101) in the study of Bassetti et al.14 to 14.3% (3 patients out of 21) in in the study of Haidar et al.,6 and around 10% in other of studies: 10.6% (5 out of 47) in Fraile-Ribot et al.,5 10.5% (4 out of 38) in Escolà-Vergé et al.15 In our study, ceftolozane/tazobactam resistance emerged in 10.2% of the treated patients (5 patients out of 49). Resistance development is expected to occur more likely in MDR and XDR isolates (like the widespread high risk clone ST175 found in our study), as they already present one of the required mutations (that leading to AmpC overexpression)16 and therefore only one more (leading to AmpC structure modification) mutation is needed for the development of resistance.5,12
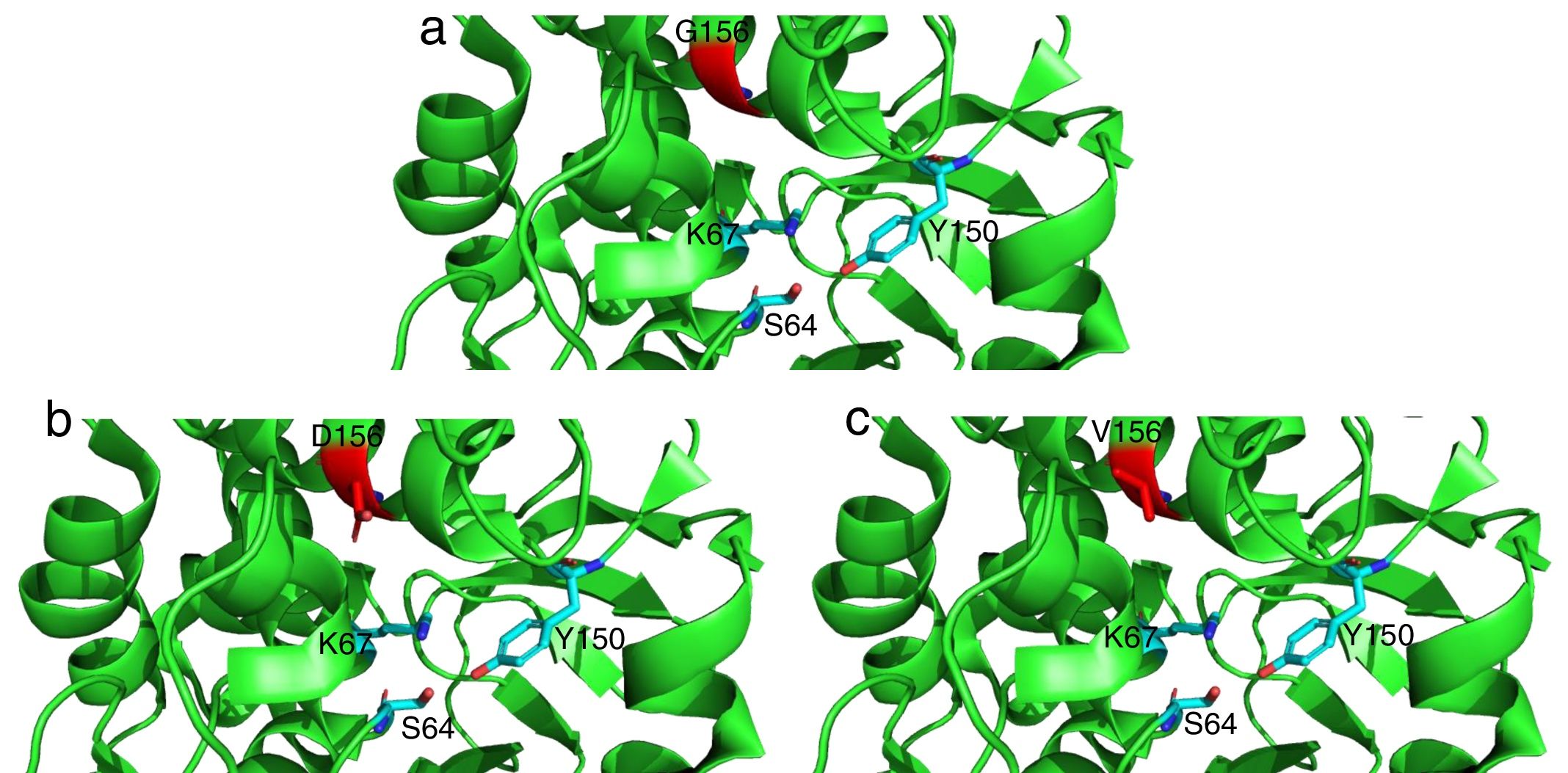
The mutations described here (with the exception of G183V mutation) have been already linked to ceftolozane/tazobactam resistance in previous studies.5,12 On the other hand, the G183V mutation has led to the emergence of a novel PDC variant (PDC-388). Moreover, previous works have demonstrated the implication of another mutation in the same residue (G183D) in ceftolozane/tazobactam resistant isolates.12,17 Recent structural modeling shows that G183 is located in the catalytic pocked and that it is likely relevant for ceftolozane and ceftazidime accommodation.18 Indeed, structural modeling (Fig. 1) shows that G183D and G183V confer a similar modification of the catalytic pocket. Likewise, previous studies have demonstrated that ceftolozane/tazobactam resistance was mediated by AmpC overexpression and mutations within the AmpC Ω loop.5,12,19 E247 is located within the Ω loop and T96 interacts directly with E247.5 These substitutions in the Ω loop (part of the active site of the enzyme) have been shown to compromise the substrate binding site and these changes are thought to impact both the catalytic efficiency and spectrum of substrate specificity of AmpC to β-lactams.5
Detailed representation of the area surrounding the active site of AmpC from P. aeruginosa PAO1 (light green, PDB code 4ZDB) using the PyMol Molecular Graphic System, v.1.8 (www.pymol.com). Residues in position 183 (156 according to recent consensus numbering of AmpC β-lactamases23) are colored in red: (a) G156, (b) D156 and (c) V156. Active residues (S64, K67 and Y150) are indicated as well.
Moreover, the acquisition of exogenous β-lactamase enzymes is another growing concern in the development of ceftolozane/tazobactam resistance, especially in the high-risk clones. These resistance mechanisms are often associated with class 1 integrons which increase the potential of dissemination.9,20,21 Recent reports have described the development of resistance to ceftolozane/tazobactam and ceftazidime/avibactam through the selection of extended-spectrum mutations from narrow spectrum OXA β-lactamases (OXA-2 or OXA-10).9,22 However, all five pairs of isolates tested in this study were negative for OXA-2 or OXA-10 β-lactamases, ruling out this possibility. Furthermore, there are two reports related to ceftolozane/tazobactam resistance development through acquisition of specific extended-spectrum β-lactamases (ESBL). Poirel et al.20 reported a P. aeruginosa MDR isolate belonging to ST235 which produced a GES-6 enzyme. This variant was not only an ESBL; the resistance profile also included carbapenems and ceftolozane/tazobactam. Khan et al.21 reported two ST309 P. aeruginosa isolates which harbored the ESBL GES-19 and GES-26 variants. Interestingly, in these last reports, MICs of ceftazidime/avibactam and aztreonam remained in the susceptibility range. This recent combination might offer an effective therapeutic option for treating infections caused by P. aeruginosa MDR/XDR isolates producing GES-variants.
As described previously,5,12 in our case, the emerging ceftolozane/tazobactam resistance restored the susceptibility to imipenem and piperacillin/tazobactam. This fact could be taken into account to expand treatment options, although clinical experience is needed in this aspect to know if these agents could be used safely.
Our study has some limitations; we focused on the study of the AmpC mutations, the main ceftolozane/tazobactam resistance mechanism described until now, but a complete characterization of all the resistance mechanisms present in these isolates and the possible implication in ceftolozane/tazobactam and ceftazidime/avibactam resistance would have been desirable.
In summary, we report the AmpC β-lactamase mutations leading to ceftolozane/tazobactam emerging resistance during therapy in XDR P. aeruginosa infections, with the description of a novel PDC variant (PDC-388). All resistant isolates showed cross-resistance to ceftazidime/avibactam, which severely compromises the selection of appropriate treatments.
There is a need for surveillance and characterization of emerging ceftolozane/tazobactam resistance, in order to preserve this valuable antipseudomonal agent.
Conflicts of interestAO has received fees as speaker and research grants for MSD and Pfizer.
MF has received a grant (“Ayuda para la formación”) from the Sociedad Española de Enfermedades Infecciosas y Microbiología Clínica (SEIMC).