The analytical performance of the new Alere™ i Influenza A&B kit (AL-Flu) assay, based on isothermal nucleic acids amplification, was evaluated and compared with an antigen detection method, SD Bioline Influenza Virus Antigen Test (SDB), and an automated real-time RT-PCR, Simplexa™ Flu A/B & VRS Direct assay (SPX), for detection of influenza viruses. An “in-house” RT-PCR was used as the reference method. Sensitivity of AL-Flu, SDB, and SPX was 71.7%, 34.8%, and 100%, respectively. Specificity was 100% for all techniques. The turnaround time was 13min for AL-Flu, 15min for SDB, and 75min for SPX.
The Alere™ i Influenza A&B assay is an optimal point-of-care assay for influenza diagnosis in clinical emergency settings, and is more sensitive and specific than antigen detection methods.
Se evaluó el nuevo ensayo Alere™ i Influenza A&B kit (AL-Flu), basado en la amplificación isotérmica de ácidos nucleicos, y se comparó con un método de detección de antígeno, SD Bioline Influenza Virus Antigen Test (SDB), y con una RT-PCR en tiempo real automática, Simplexa™ Flu A/B & VRS Direct assay (SPX), para la detección de virus de la gripe. Se utilizó una RT-PCR en tiempo real casera como método de referencia. La sensibilidad de AL-Flu, SDB y SPX fue del 71,7%, del 34,8% y del 100%, respectivamente. Se obtuvo una especificidad del 100% con todos los métodos. El tiempo de realización fue de 13min para AL-Flu, de 15min para SDB y de 75min para SPX.
El ensayo Alere™ i Influenza A&B es óptimo para el diagnóstico de gripe en unidades de urgencias, al ser más sensible y específico que las técnicas de detección de antígeno.
Rapid influenza antigen detection tests (RADTs) have been recommended as point-of-care tools for influenza virus (Flu) detection; however they have shown variable sensitivity (16–83%).1,2 A negative result by RADT does not exclude infection, and requires confirmation with more sensitive methods.2 Automated nucleic acid amplification tests (NAATs) based on real-time reverse transcriptase-PCR (rRT-PCR) are currently available that, although highly sensitive, they are more expensive and require a longer turnaround time than RADTs.3–6
The new Alere™ i Influenza A&B kit (AL-Flu, Alere Scarborough, Scarborough, ME) has shown excellent sensitivity and specificity values compared with viral culture although variable performance compared with NAATs.7–11 The system uses isothermal nucleic acid amplification technology, is intended for direct specimen testing and provides results in 13min with minimal hands-on time. In this study, the analytical performance of AL-Flu was compared with an immunochromatographic RADT and an automated rRT-PCR.
MethodsA retrospective study was conducted on 70 specimens (31 nasopharyngeal aspirates, 36 nasopharyngeal swabs and 3 bronchial aspirates, conserved at −80°C) from patients with acute respiratory tract infection during the 2013–2014 period. Flu-positive samples (n=49) were previously characterized as Flu A (n=24) or Flu B (n=25) either by an “in-house” rRT-PCR (h-rRT-PCR)12 (43 specimens) or by SDB (6 specimens). Flu-negative samples (n=21) were positive to other respiratory virus by the xTAG® RVP Fast v2 kit (Luminex Diagnostics, Toronto, Canada). A total of 30 viruses were detected in these 21 Flu-negative samples: 4 adenovirus, 4 bocavirus, 1 coronavirus NL63, 8 enterovirus/rhinovirus, 4 human metapneumovirus (hMPV), 8 parainfluenza viruses (PIV) 1–4 and 1 respiratory syncytial virus.
Respiratory samples were thawed, briefly mixed and separated into four aliquots for Flu testing by AL-Flu, SD Bioline Influenza Virus Antigen Test (SDB, Alere Healthcare SLU, Barcelona, Spain), the automated Simplexa™ Flu A/B & VRS Direct assay (SPX, Focus Diagnostics, Cypress, CA), and by h-rRT-PCR, which was considered the reference method. The h-rRT-PCR was carried out as described by Bonzel et al.,12 with modified primers SW.NP-F (5′-CAG CTG CMC ARA GRG CAA TGR TG-3′) and SW.NP-R (5′-RAA DAT GAG RTC TTC RAT YTC AGC-3′), and Taqman® probe SW.NP-pr (5′-6FAM – CAA GTR AGA GAA AGY CGR AAC CCA GG-BHQ1-3′). The threshold cycle value (Ct) of the h-rRT-PCR was recorded in order to semi-quantify the viral load.
Flu A-positive samples were subsequently subtyped as H1pdm09 (n=14) or H3 (n=10) as previously described,13 with modified primers SWH1.737-F (5′-ATT ACT GGA CAC TAG TAG AG-3) and SWH1.833-R (5′-GCA TTT CTT TCC ATT GCG AAT G-3′), and Taqman® probe SWH1.808-pr (5′-6FAM-TCG AAG CAA CTG GAA ATC TAG TGG TAC CG-BHQ1-3′).
The study protocol was carried out in accordance with the Declaration of Helsinki. This was a non-interventional study with no additional investigation to routine procedures. No additional sampling or modification to the physician prescription was performed. Data analyses were carried out using an anonymous database.
Data were analyzed with SPSS 15.0.1 software (SPSS Inc., Chicago, IL). Sensitivity, specificity, positive (PPV) and negative predictive values (NPV) with their 95% confidence intervals (CI) of all methods were determined. Two-by-two concordances were estimated by the Cohen's kappa coefficient of agreement (k). Mean Ct values of the h-rRT-PCR of positive and false-negative samples by each method were compared with the Mann Whitney U test. A p value <0.05 was considered significant.
ResultsThe h-rRT-PCR, carried out in thawed samples, was positive to Flu viruses in 46 (true positives) and negative in 3 out of the 49 previously positives. The Ct (mean±standard deviation) was 27.9±4.23.
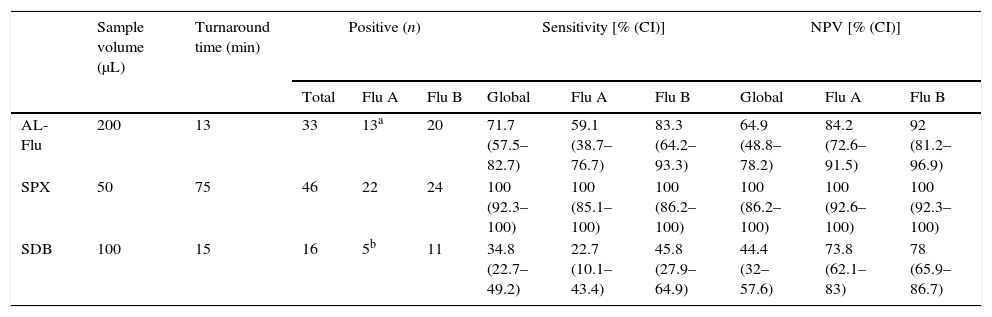
The specificity and PPV of all tests was 100%. No false positive results were obtained by any method, and Flu types were correctly classified in all positive samples. Indeed, the 3 negative samples by h-rRT-PCR, previously positives in routine, were also negative with AL-Flu, SDB and SPX. Sensitivity and NPV of AL-Flu, SDB and SPX were 71.7% and 64.9%, 34.8% and 44.4%, and 100% and 100%, respectively (Table 1).
Analytical performance of Alere™ i Influenza A&B kit (AL-Flu), Simplexa™ Flu A/B&VRS Direct assay (SPX) and SD Bioline Influenza Virus Antigen Test (SDB).
| Sample volume (μL) | Turnaround time (min) | Positive (n) | Sensitivity [% (CI)] | NPV [% (CI)] | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Flu A | Flu B | Global | Flu A | Flu B | Global | Flu A | Flu B | |||
| AL-Flu | 200 | 13 | 33 | 13a | 20 | 71.7 (57.5–82.7) | 59.1 (38.7–76.7) | 83.3 (64.2–93.3) | 64.9 (48.8–78.2) | 84.2 (72.6–91.5) | 92 (81.2–96.9) |
| SPX | 50 | 75 | 46 | 22 | 24 | 100 (92.3–100) | 100 (85.1–100) | 100 (86.2–100) | 100 (86.2–100) | 100 (92.6–100) | 100 (92.3–100) |
| SDB | 100 | 15 | 16 | 5b | 11 | 34.8 (22.7–49.2) | 22.7 (10.1–43.4) | 45.8 (27.9–64.9) | 44.4 (32–57.6) | 73.8 (62.1–83) | 78 (65.9–86.7) |
CI, 95% confidence interval; NPV, negative predictive value.
The Ct (mean±standard deviation) of positive samples by AL-Flu and SDB were 26.21±4.04 and 23.26±2.16, respectively. The Ct (mean±standard deviation) of false negative samples by AL-Flu and SDB were 31.4±1.81 and 29.45±3.57, respectively. Significant differences in mean Ct values between positive and negative samples by AL-Flu an SDB were observed (p<0.001). A good agreement (k=0.635) was observed between AL-Flu and SPX, whereas moderate (k=0.499) and weak (k=0.268) agreements were obtained comparing SDB with AL-Flu and SPX, respectively.
Total turnaround time of AL-Flu, SDB and SPX was 13, 15 and 75min, respectively.
DiscussionAL-Flu showed a significantly higher sensitivity than SDB for Flu detection, both requiring the same turnaround time. The superior performance of AL-Flu compared with other RADTs has been previously reported, with similar results.7,9 Recently, new RADTs have demonstrated higher sensitivity than immunochromatographic assays and similar to the one demonstrated for AL-Flu in this study.2
However, we obtained a lower sensitivity of AL-Flu than previous reports, comparing the assay with NAATs based on PCR.8,10,11 It may reflect variable viral amount in the sample among all studies. In this study, we semi-quantified the viral load by the Ct measure of the h-rRT-PCR. Mean Ct was higher than the one reported in samples tested by Hurtado et al.,14 who demonstrated a better positive percent agreement with a h-rRT-PCR. Although results from both studies may not be comparable since our h-rRT-PCR, used as the reference method, is probably different to the one used by Hurtado et al., we observed significant differences in mean Ct between positive and negative samples by AL-Flu. By our results, samples with a Ct >30 in the h-rRT-PCR would likely give a false negative result. Although high viral loads are expected in patients with Flu if laboratory diagnosis is performed within 48–72h after the onset of symptoms (http://www.cdc.gov/flu/professionals/diagnosis/molecular-assays.htm), delayed sampling is feasible, especially during inter-epidemic periods. In these situations, a highly sensitive method is necessary to achieve a reliable diagnosis.
On the contrary, this study demonstrated a 100% specificity in contrast to those reported by other investigators who found false positives or Flu typing errors.8,11
Cost-effective diagnostic algorithms should be established, based on clinical features and laboratory resources. Once Flu circulation is documented in a geographical region, a clinical diagnosis could be enough for outpatients. In these situations, Flu testing should only be performed in special situations (severe cases and/or high-risk conditions) (http://www.cdc.gov/flu/professionals/diagnosis/clinician_guidance_ridt.htm).
SPX and AL-Flu demonstrated higher sensitivity than SDB. Due to its speed and ease of use, AL-Flu may be an optimal alternative to RADTs as a point-of-care method for Flu detection in clinical emergency settings. However, a negative result should be confirmed with a more sensitive assay in hospitalized patients to ensure an appropriate management and treatment.
Conflict of interestThe authors have no conflict of interest to declare.