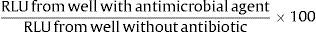The antibiotic susceptibility test (AST) in Clinical Microbiology laboratories is still time-consuming, and most procedures take 24h to yield results. In this study, a rapid antimicrobial susceptibility test using ATP-bioluminescence has been developed. The design of method was performed using five ATCC collection strains of known susceptibility. This procedure was then validated against standard commercial methods on 10 strains of enterococci, 10 staphylococci, 10 non-fermenting gram negative bacilli, and 13 Enterobacteriaceae from patients. The agreement obtained in the sensitivity between the ATP-bioluminescence method and commercial methods (E-test, MicroScan and VITEK2) was 100%. In summary, the preliminary results obtained in this work show that the ATP-bioluminescence method could provide a fast and reliable AST in two hours.
La mayoría de procedimientos diagnósticos para el estudio de la sensibilidad de las bacterias a los antibióticos en Microbiología Clínica requieren unas 24 horas para la obtención de resultados. En este estudio se propone una metodología para llevar a cabo un antibiograma rápido mediante la medición de ATP por bioluminiscencia. El diseño del antibiograma se realizó mediante el uso de cinco cepas de colección ATCC, las cuales presentan una sensibilidad conocida. Este diseño fue posteriormente validado frente a los métodos comerciales de antibiograma mediante el procesamiento de 10 cepas de enterococos, 10 de estafilococos, 10 de bacilos gramnegativos no fermentadores y 13 de Enterobacteriaceae aisladas de pacientes. El acuerdo obtenido entre la sensibilidad obtenida mediante bioluminiscencia y la obtenida mediante los métodos comerciales (E-test, MicroScan and VITEK2) fue del 100%. Por lo tanto, los resultados preliminares obtenidos en este trabajo indican que las medidas de ATP mediante bioluminiscencia podrían proporcionar, en dos horas, un antibiograma rápido y seguro.
The number of patients who suffer bacterial infections is increasing due to the increase in life expectancy and aggressive medical treatments. Parallel to this, the emergence of numerous multidrug resistance pathogenic strains is well documented.1 Taking into account that the early administration of the correct antibiotic increases survival of patients2 and that most of the antibiotic susceptibility tests (AST) in Clinical Microbiology take 24h,3 there is a need for achieving a faster and accurate AST. For this reason, various methodologies, such as flow cytometry, molecular detection techniques, bioluminescence, chemiluminescence, nephelometry, microarrays or colourimetric methods have been introduced in Clinical Microbiology laboratories.
Bioluminescence is the production and emission of light by an organism as the result of a chemical reaction during which chemical energy is converted into radiant energy. Firefly bioluminescent ATP assay, by means of the luciferin–luciferase reaction and in the presence of ATP, releases light.4 ATP is a universal metabolite in living cells and the light produced is proportional to the amount of bacterial ATP present in the sample, and hence proportional to the bacterial cells. Therefore, this assay allows indirectly quantifying bacterial cells number.5 When firefly bioluminescent ATP assay has been applied to AST, the time required to achieve results ranged from four to six hours. Moreover, non-fermenting gramnegative rods (NFGNR) still remain to be tested.5–7 In this context, the aim of this work was to perform a rapid and complete AST using a firefly bioluminescent ATP assay in which the most predominant bacteria isolated in Clinical Microbiology laboratories were tested against some used antibiotics in clinical practice.
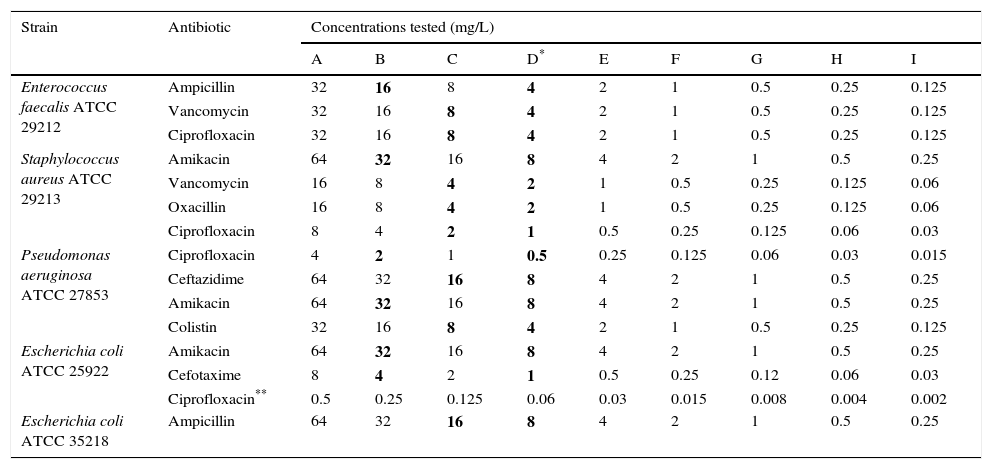
Materials and methodsExperimental design of the antimicrobial susceptibility testStandard antimicrobial powder of Cefotaxime, Amikacin, Ciprofloxacin, Vancomycin, Oxacillin, Colistin, Ceftazidime and Ampicillin were obtained from Sigma-Aldrich (St. Louis, MO, USA). Stock solutions were prepared according to the protocol proposed by the EUCAST,8 sterilized by filtration using a MILLEX GS 0.22μm membrane filter (EMD® Millipore Corporation, Billerica, MA, USA) and stored at −80°C. Table 1 summarizes the ATCC collection strains and the antibiotics used to set up the proposed AST, which was performed in a 96-well white opaque microtiter plate with an F-shaped bottom (Thermo Scientific, Waltham, MA, USA). The concentrations of antibiotic tested were achieved by serial dilutions. The medium used was cation-adjusted Mueller-Hinton broth (CAMHB) (Difco, Sparks, MD, USA). 1.3μl of 0.5-McFarland-standard bacterial suspension were added into wells; in this way, the final inoculum was approximately 9.7×105CFU/ml. For Oxacillin the medium was supplemented with 2% NaCl. Moreover, for each strain tested, a well without antibiotic was prepared adding 200μl of CAMHB and 1.3μl of 0.5-McFarland-standard bacterial suspension.
Bacterial strains and concentrations of the antibiotics used to set up the proposed method.
| Strain | Antibiotic | Concentrations tested (mg/L) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D* | E | F | G | H | I | ||
| Enterococcus faecalis ATCC 29212 | Ampicillin | 32 | 16 | 8 | 4 | 2 | 1 | 0.5 | 0.25 | 0.125 |
| Vancomycin | 32 | 16 | 8 | 4 | 2 | 1 | 0.5 | 0.25 | 0.125 | |
| Ciprofloxacin | 32 | 16 | 8 | 4 | 2 | 1 | 0.5 | 0.25 | 0.125 | |
| Staphylococcus aureus ATCC 29213 | Amikacin | 64 | 32 | 16 | 8 | 4 | 2 | 1 | 0.5 | 0.25 |
| Vancomycin | 16 | 8 | 4 | 2 | 1 | 0.5 | 0.25 | 0.125 | 0.06 | |
| Oxacillin | 16 | 8 | 4 | 2 | 1 | 0.5 | 0.25 | 0.125 | 0.06 | |
| Ciprofloxacin | 8 | 4 | 2 | 1 | 0.5 | 0.25 | 0.125 | 0.06 | 0.03 | |
| Pseudomonas aeruginosa ATCC 27853 | Ciprofloxacin | 4 | 2 | 1 | 0.5 | 0.25 | 0.125 | 0.06 | 0.03 | 0.015 |
| Ceftazidime | 64 | 32 | 16 | 8 | 4 | 2 | 1 | 0.5 | 0.25 | |
| Amikacin | 64 | 32 | 16 | 8 | 4 | 2 | 1 | 0.5 | 0.25 | |
| Colistin | 32 | 16 | 8 | 4 | 2 | 1 | 0.5 | 0.25 | 0.125 | |
| Escherichia coli ATCC 25922 | Amikacin | 64 | 32 | 16 | 8 | 4 | 2 | 1 | 0.5 | 0.25 |
| Cefotaxime | 8 | 4 | 2 | 1 | 0.5 | 0.25 | 0.12 | 0.06 | 0.03 | |
| Ciprofloxacin** | 0.5 | 0.25 | 0.125 | 0.06 | 0.03 | 0.015 | 0.008 | 0.004 | 0.002 | |
| Escherichia coli ATCC 35218 | Ampicillin | 64 | 32 | 16 | 8 | 4 | 2 | 1 | 0.5 | 0.25 |
For performing luminometric measurement, microplates were introduced into the luminometer (Appliskan, Thermo Scientific, Waltham, MA, USA). After aerobic incubation for two hours at 35°C, ATP was extracted from microbial cultures and accurately measured as follows: 20μl of ATP extractant mixture (BioThema AB, Handen, Sweden) was added using the dispenser of the instrument, the microplate was shaken with an amplitude of 5mm for 15s and 100μl of ATP reagent HS (BioThema AB, Handen, Sweden) was added with the dispenser. The measurement mode was high sensitivity for 2000ms. Ultimately, the software (Thermo Scientific SkanIt) provided us the relative light units (RLU) produced as a result of ATP hydrolysis in each of these steps, which are proportional to ATP concentration. ATP-bioluminescence obtained from each well was expressed as a percentage with the formula:
Finally, the percentages obtained were analyzed by receiver operator characteristic (ROC) curves to determine the optimal threshold for discriminating between susceptibility and resistance.
In order to evaluate the precision of ATP-bioluminescence signals, the AST performed on ATCC collection strains was repeated in triplicate the same working session and one per day for three days.
Validation procedureThe procedure was validated with 43 clinical strains identified from colonies obtained in culture plates using MALDI-TOF (Bruker Daltonik GmbH, Bremen, Germany). In enterococci, seven Enterococcus faecalis and three Enterococcus faecium were tested against Ampicillin, Vancomycin and Ciprofloxacin at concentrations of 32, 16, 8, 4, 2 and 1mg/l. In staphylococci, four Staphylococcus aureus, two Staphylococcus epidermidis, two Staphylococcus hominis and one Staphylococcus haemolyticus and Staphylococcus lugdunensis were included. These strains were tested against Amikan and Ciprofloxacin at 64, 32, 16, 8, 4 and 2mg/l and 8, 4, 2, 1, 0.5 and 0.25mg/l respectively; for Vancomycin, S. aureus were tested at 16, 8, 4, 2, 1 and 0.5mg/l, and coagulase negative staphylococci (CNS) and S. lugdunensis were tested at 32, 16, 8, 4, 2 and 1mg/l; and for Oxacillin, S. aureus and S. lugdunensis were tested at 16, 8, 4, 2, 1 and 0.5mg/l, and CNS at 2, 1, 0.5, 0.25, 0.125 and 0.0625mg/l. In Enterobacteriaceae, four Proteus mirabilis, two Escherichia coli and one Klebsiella pneumoniae, Klebsiella oxytoca, Serratia marcescens, Providencia stuartii, Enterobacter cloacae, Citrobacter freundii and Enterobacter aerogenes were tested against Ampicillin and Amikacin at 64, 32, 16, 8, 4 and 2mg/l; Cefotaxime at 8, 4, 2, 1, 0.5 and 0.25mg/l; and Ciprofloxacin at 4, 2, 1, 0.5, 0.25 and 0.125mg/l. In NFGNR, three Pseudomonas aeruginosa and one Pseudomonas stutzeri and Pseudomonas putida were tested against Ceftazidime and Amikacin at 64, 32, 16, 8, 4 and 2mg/l; Colistin at 32, 16, 8, 4, 2 and 1mg/l; and Ciprofloxacin at 4, 2, 1, 0.5, 0.25 and 0.125mg/l. Three Acinetobacter baumannii and one Acinetobacter ursingii and Acinetobacter johnsonii were tested against Amikacin at 64, 32, 16, 8, 4 and 2mg/l; Colistin at 16, 8, 4, 2, 1 and 0.5mg/l; and Ciprofloxacin at 8, 4, 2, 1, 0.5 and 0.25mg/l.
The results of susceptibility obtained at 35°C from colonies grown in culture plates by means of the commercial methods VITEK2 (bioMérieux, Marcy l’Etoile, France), MicroScan (Siemens, Tarrytown, NY, USA) and E-test (Liofilchem, Roseto Degli Abruzzi, Italy), applying the criteria published by the EUCAST,9 were considered as the gold standard. Finally, results obtained by ATP-bioluminescence were compared to the gold standard according to FDA criteria10; agreements and disagreements among the susceptibility values obtained were classified as agreements, very major errors (false susceptibility), major errors (false resistance), or minor errors (susceptible/resistant versus intermediate susceptibility). Moreover, Kappa concordance index was calculated to analyze the degree of disagreement with the gold standard test.
All the statistical calculations were performed using the SPSS v.20.0 software.
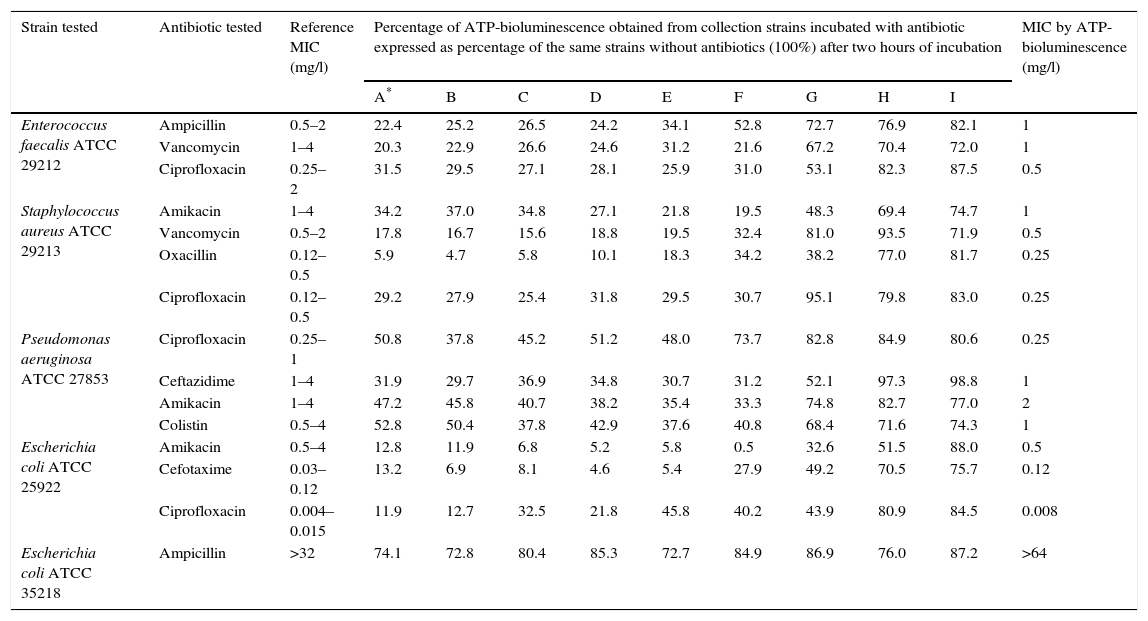
ResultsTable 2 summarizes the results obtained from ATCC collection strains incubated with antibiotic expressed as percentage of the ATP-bioluminescence of the same strains incubated without antibiotics (100%). In order to obtain the optimal cut-off ATP-bioluminescence for discriminating between sensitivity and resistance, data represented in Table 2 were analyzed by ROC curve. Table 3 shows a complete sensitivity/specificity report and indicates the cut-off value in bold. This value allowed us to consider a bacterium sensitive to the antibiotic tested if a ATP-bioluminescence reduction of at least 36.8% in the presence of such antibiotic at a concentration less than or equal to the stated breakpoint concentration for sensitivity compared to the signal obtained from the same strain incubated without antibiotic was obtained after two hours of incubation. Otherwise, a bacterium was considered to be resistant to the antibiotic tested if in order to obtain a ATP-bioluminescence reduction of at least 36.8% in the presence of such antibiotic after two hours of incubation compared to the same strain without added antibiotic, the concentration of antibiotic required is higher than or equal to the stated breakpoint concentration for resistance. Applying this cut-off, a 100% concordance between the results of susceptibility obtained by ATP-bioluminescence and by the commercial methods (VITEK2, MicroScan and E-test) was observed in the ATCC collection strains used in the design of the proposed method (Table 2).
Results obtained from ATCC collection strains by ATP-bioluminescence.
| Strain tested | Antibiotic tested | Reference MIC (mg/l) | Percentage of ATP-bioluminescence obtained from collection strains incubated with antibiotic expressed as percentage of the same strains without antibiotics (100%) after two hours of incubation | MIC by ATP-bioluminescence (mg/l) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A* | B | C | D | E | F | G | H | I | ||||
| Enterococcus faecalis ATCC 29212 | Ampicillin | 0.5–2 | 22.4 | 25.2 | 26.5 | 24.2 | 34.1 | 52.8 | 72.7 | 76.9 | 82.1 | 1 |
| Vancomycin | 1–4 | 20.3 | 22.9 | 26.6 | 24.6 | 31.2 | 21.6 | 67.2 | 70.4 | 72.0 | 1 | |
| Ciprofloxacin | 0.25–2 | 31.5 | 29.5 | 27.1 | 28.1 | 25.9 | 31.0 | 53.1 | 82.3 | 87.5 | 0.5 | |
| Staphylococcus aureus ATCC 29213 | Amikacin | 1–4 | 34.2 | 37.0 | 34.8 | 27.1 | 21.8 | 19.5 | 48.3 | 69.4 | 74.7 | 1 |
| Vancomycin | 0.5–2 | 17.8 | 16.7 | 15.6 | 18.8 | 19.5 | 32.4 | 81.0 | 93.5 | 71.9 | 0.5 | |
| Oxacillin | 0.12–0.5 | 5.9 | 4.7 | 5.8 | 10.1 | 18.3 | 34.2 | 38.2 | 77.0 | 81.7 | 0.25 | |
| Ciprofloxacin | 0.12–0.5 | 29.2 | 27.9 | 25.4 | 31.8 | 29.5 | 30.7 | 95.1 | 79.8 | 83.0 | 0.25 | |
| Pseudomonas aeruginosa ATCC 27853 | Ciprofloxacin | 0.25–1 | 50.8 | 37.8 | 45.2 | 51.2 | 48.0 | 73.7 | 82.8 | 84.9 | 80.6 | 0.25 |
| Ceftazidime | 1–4 | 31.9 | 29.7 | 36.9 | 34.8 | 30.7 | 31.2 | 52.1 | 97.3 | 98.8 | 1 | |
| Amikacin | 1–4 | 47.2 | 45.8 | 40.7 | 38.2 | 35.4 | 33.3 | 74.8 | 82.7 | 77.0 | 2 | |
| Colistin | 0.5–4 | 52.8 | 50.4 | 37.8 | 42.9 | 37.6 | 40.8 | 68.4 | 71.6 | 74.3 | 1 | |
| Escherichia coli ATCC 25922 | Amikacin | 0.5–4 | 12.8 | 11.9 | 6.8 | 5.2 | 5.8 | 0.5 | 32.6 | 51.5 | 88.0 | 0.5 |
| Cefotaxime | 0.03–0.12 | 13.2 | 6.9 | 8.1 | 4.6 | 5.4 | 27.9 | 49.2 | 70.5 | 75.7 | 0.12 | |
| Ciprofloxacin | 0.004–0.015 | 11.9 | 12.7 | 32.5 | 21.8 | 45.8 | 40.2 | 43.9 | 80.9 | 84.5 | 0.008 | |
| Escherichia coli ATCC 35218 | Ampicillin | >32 | 74.1 | 72.8 | 80.4 | 85.3 | 72.7 | 84.9 | 86.9 | 76.0 | 87.2 | >64 |
Coordinates of the ROC curve for discriminating between sensitivity and resistance calculated by SPSS v.20.0 software.
| Hypothetical percentage of ATP-bioluminescence expressed as the percentage of the controls without added antibiotics (100%) | Sensitivity | Specificity |
|---|---|---|
| 47.600 | 1.000 | 0.084 |
| 49.200 | 1.000 | 0.072 |
| 50.600 | 1.000 | 0.060 |
| 51.000 | 1.000 | 0.048 |
| 52.000 | 1.000 | 0.036 |
| 53.250 | 1.000 | 0.012 |
| 63.200 | 1.000 | 0.000 |
| 72.750 | 0.833 | 0.000 |
| 73.450 | 0.667 | 0.000 |
| 77.250 | 0.500 | 0.000 |
In bold is indicated the cut-off percentage of ATP-bioluminescence which allows to discriminate between strains susceptibility and resistance to antibiotics with 100% sensitivity and 100% specificty.
When the precision of the signals of ATP-bioluminiscence was evaluated, the intra-day and inter-day precision, expressed as relative standard deviation, were in the ranges 5–12 and 9–18%, respectively.
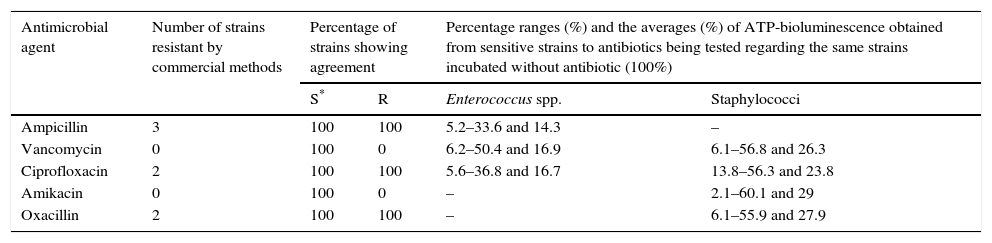
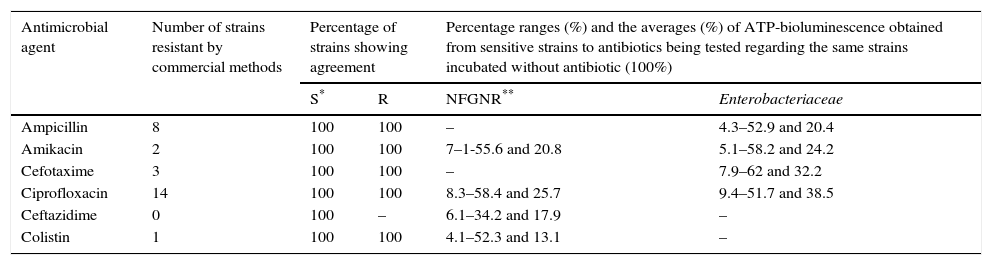
For validation of the proposed AST against commercial methods, 43 clinical strains isolated at University Clinic Hospital of Valladolid were processed. The results of the susceptibility tests performed by commercial methods (VITEK 2, MicroScan and E-test) from colonies obtained in the culture showed 100% concordance among them. The AST performed by ATP-bioluminescence from 10 enterococci, 10 staphylococci, 10 NFGNR and 13 Enterobacteriaceae showed 100% concordance with the susceptibility results obtained by the commercial methods (Tables 4 and 5). In this way, neither major errors nor minor errors were obtained and the Kappa index calculation provided a value of 0.9688 (IC 95%: 0.9126–1.0000) (p<0.001).
Results obtained from 20 clinical strains of grampositive bacteria.
| Antimicrobial agent | Number of strains resistant by commercial methods | Percentage of strains showing agreement | Percentage ranges (%) and the averages (%) of ATP-bioluminescence obtained from sensitive strains to antibiotics being tested regarding the same strains incubated without antibiotic (100%) | ||
|---|---|---|---|---|---|
| S* | R | Enterococcus spp. | Staphylococci | ||
| Ampicillin | 3 | 100 | 100 | 5.2–33.6 and 14.3 | – |
| Vancomycin | 0 | 100 | 0 | 6.2–50.4 and 16.9 | 6.1–56.8 and 26.3 |
| Ciprofloxacin | 2 | 100 | 100 | 5.6–36.8 and 16.7 | 13.8–56.3 and 23.8 |
| Amikacin | 0 | 100 | 0 | – | 2.1–60.1 and 29 |
| Oxacillin | 2 | 100 | 100 | – | 6.1–55.9 and 27.9 |
Results obtained from 23 clinical strains of gramnegative bacteria.
| Antimicrobial agent | Number of strains resistant by commercial methods | Percentage of strains showing agreement | Percentage ranges (%) and the averages (%) of ATP-bioluminescence obtained from sensitive strains to antibiotics being tested regarding the same strains incubated without antibiotic (100%) | ||
|---|---|---|---|---|---|
| S* | R | NFGNR** | Enterobacteriaceae | ||
| Ampicillin | 8 | 100 | 100 | – | 4.3–52.9 and 20.4 |
| Amikacin | 2 | 100 | 100 | 7–1-55.6 and 20.8 | 5.1–58.2 and 24.2 |
| Cefotaxime | 3 | 100 | 100 | – | 7.9–62 and 32.2 |
| Ciprofloxacin | 14 | 100 | 100 | 8.3–58.4 and 25.7 | 9.4–51.7 and 38.5 |
| Ceftazidime | 0 | 100 | – | 6.1–34.2 and 17.9 | – |
| Colistin | 1 | 100 | 100 | 4.1–52.3 and 13.1 | – |
In the 1980s appeared some instrumental techniques in Clinical Microbiology laboratories, such as bioluminescence and flow cytometry, which allow obtaining a rapid AST.11 Nevertheless, these techniques have a major drawback: bacterial identification cannot be obtained. For this reason these techniques were not implemented given that for MIC interpretation it is indispensable to know the bacterial species being studied. Thus, when these techniques were evaluated it was mandatory to perform an incubation of the colonies grown in culture plates for 24h in order to obtain bacterial identification.
Matrix-Assisted Laser Desorption/Ionization-time of flight mass spectrometry (MALDI-TOF-MS) allows obtaining a rapid bacterial identification (in minutes) from colonies grown in culture plates. Besides, bacterial identification can be achieved from positive blood culture bottles in one hour after the system of incubation flagged one as positive.12 Therefore, the implementation of MALDI-TOF permits the use of techniques that only perform a rapid AST. In this way, we have a good opportunity to re-examine the current techniques that provide a rapid AST in order to investigate if anyone provides an adequate sensitivity and specificity for clinical practice. In this work we have decided to re-examine the use of ATP-bioluminescence for performing a rapid AST because of its previous published applications in Clinical Microbiology, such as rapid AST in bacteria (including mycobacteria) and yeasts,4,13–15 screening diagnostic in urinalysis and bacterial detection directly from urine samples16 and bronchoalveolar lavage.17
In order to set up the proposed AST based on ATP-biolumeniscence, the RLU obtained from ATCC collection strains incubated with antibiotic were compared to the RLU obtained from the same strains incubated without antibiotic after two hours of incubation. If intracellular ATP is measured, sensitive strains to the antibiotic tested should provide RLU values lower than the same strains incubated without antibiotic because ATP is associated with living cells. This fact was not observed in ATCC collection strains after one hour of incubation (data not shown). For this reason it was not possible to achieve results in one hour. Nevertheless, after two hours of incubation it was possible to discriminate between sensitivity and resistance by comparing RLU values, and we pointed out that a cut-off value of 36.8% ATP-bioluminescence of a strain incubated with antibiotic compared to the same strain incubated without antibiotic provided a 100% specificity and 100% sensitivity to differentiate between sensitive and resistance.
With regard to the precision of ATP-bioluminescence signals, the inter-day relative standard deviation was higher than that intra-day. This fact was possibly due to that inter-day experiments were performed using different 0.5-McFarland-standard bacterial suspensions. Taking into account that turbidimetry is not an accurate method for preparing bacterial suspensions, initial bacterial concentrations achieved in the same wells were different. In this way, the ATP detected from the same wells after two-hour incubation was different. On the other hand, the intra-day AST were performed using the same 0.5-McFarland-standard bacterial suspension and, hence, the ATP detected from the same wells after two-hour incubation was more accurate.
The AST by ATP-bioluminescence proposed required two hours of incubation to achieve satisfactory results and the time previously reported for the same determination ranged from four to six hours.5–7 Given that a reduction of the time needed to perform an AST has been achieved, our method would improve patients’ outcome,2 could decrease the health care expenditure18 and may delay the rise of bacterial resistances.1 Wheat et al.7 tested 76 clinical isolates of Enterobacteriaceae to Ampicillin, Piperacillin and Gentamicin by ATP bioluminescence. They obtained satisfactory results for most organisms tested, except for P. mirabilis. In order to achieve better results for P. mirabilis, a reduction of the osmolality broth by adding distilled water was carried out. In this way, the lysis of spheroplasts took place and a much better correlation between ATP susceptibility results and standard MIC values was achieved for P. mirabilis. Our results for P. mirabilis correlated 100% with the susceptibility obtained from commercial methods and improved those obtained by Wheat et al.7 This fact could be explained by some reasons. One of them might be that the osmolality of the cation-adjusted Mueller-Hinton broth is different from the Isosensitest broth. But, the most important point is possibly that we measured the ATP present in 200μl of sample and we diluted all reagents at 1/16. Otherwise, Wheat et al.7 measured the ATP present in only 20μl of sample and diluted reagents at 1/52. When this higher dilution is performed it is possible that the ATP Monitoring Reagent loses its ability to produce bacterial lysis. Thus, ATP measurements were only reliable when data were obtained from strains of P. mirabilis incubated in a medium with low osmolality, in which bacterial lyses occurs more easily. Moreover, it should be noted that the studies were performed with different reagents, experimental conditions and equipments.
Wheat et al.6 tested the susceptibility of S. aureus and CNS to Oxacillin at 30°C and 37°C, respectively, and 6% of major errors were obtained. Our results were better than those obtained by Wheat et al.6 probably due to the aforementioned points for P. mirabilis. Besides, we previously established the optimal cut-off ATP-bioluminescence for discriminating between sensitivity and resistance from data obtained of ATCC collection strains. This point enhances the validity of the proposed AST given that these strains show a known susceptibility and they are commonly used as quality control for AST. Wheat et al.6 when tested grampositive bacteria, did not apply any criterion previously established from data obtained of ATCC collection strains and categorized the results obtained empirically.
According to EUCAST8, Oxacillin susceptibility test on staphylococci should be performed at 30°C. Nevertheless, when we performed the test at 30°C, we observed that sensitive strains of CNS to Oxacillin incubated without Oxacillin provided an ATP signal close to that obtained from the same strains incubated with Oxacillin. In this way, a false resistance was obtained for the majority of the strains tested. However, when the same test was performed at 35°C, the sensitive strains to Oxacillin incubated without Oxacillin provided a higher ATP signal than that obtained from the same strains incubated with Oxacillin. Thus, the AST on staphylococci to Oxacillin was performed at 35°C. Besides, commercial methods used in Clinical Microbiology laboratories test the susceptibility of staphylococci to Oxacillin at 35°C.
To the best of our knowledge, this is the first time that the susceptibility to antibiotics on NFGNR has been analyzed by ATP-bioluminescence. And we demonstrated that it is possible to obtain the susceptibility on this bacterial group in two hours. Moreover, the results obtained indicated that for sensitive strains of Pseudomonas spp. to antibiotics being tested, Colistin reduced the percentage of ATP-bioluminescence more than other antibiotics. The same effect was observed for sensitive strains of Acinetobacter spp. Therefore, the most effective antibiotic tested in terms of its ability to reduce the production of bacterial ATP was Colistin. These results correlated with its fast bactericidal effect previously described in the literature.19
In the proposed AST, for ATCC collection strains, nine concentrations of antibiotic were tested (the breakpoint concentrations of sensitivity and resistance, and the concentrations in the MICs range of the ATCC collection strains). In this way, we assured that the cut-off ATP-bioluminescence obtained for discriminating between susceptibility and resistance was valid given that susceptibility and MICs obtained corresponded to those expected. For clinical strains, we tested six concentrations of antibiotic (the breakpoint concentrations of sensitivity and resistance and the nearest concentrations) because the aim of the work was to determine the susceptibility of bacteria to antibiotics by ATP-bioluminescence. Otherwise, when MICs by ATP-bioluminescence and by E-test could be compared, we observed differences in 10 cases. These differences, however, were into the range of the accepted error of the method for performing an AST, which is to say, these differences were, in all cases, of one dilution of concentration of antibiotic tested, and the interpretation of the susceptibility was the same by both methods.
The most important advantage of ATP-bioluminescence in AST is the limit detection of the method, which may be as low as 10×10−18mol ATP; this limit of detection corresponds to five bacterial cells. For this reason, it has been possible to perform an AST using a volume of only 200μl of medium. Moreover, the proposed method is easy to perform since all reagents can be dispensed automatically and hence automation of the assay for routine use is possible. On the other hand, ATP-bioluminescence encompasses some disadvantages. After mixing ATP reagents with sample the light intensity decays at approximately 6% per minute due to the chemical instability of ATP; for this reason, in this work the wells were measured singly after adding reagents. Moreover, reagents, once reconstituted, cannot be stored for more than a week. However, the cost of luciferin–luciferase reagents is the strongest argument against the use of ATP-based methods in routine diagnostic laboratories. In our study, the measurement of a 96-well microtiter plate has cost approximately 125–130€. This is an unaffordable cost for any laboratory due to the large number of AST performed daily.
Nevertheless, taken into account that ATP-bioluminescence allows obtaining the susceptibility of the main bacteria isolated in the laboratory, lowering costs of reagents, selecting bacterial strains isolated from severe patients and testing more antibiotics, this technique could be implemented in work routine of Clinical Microbiology laboratories.
In conclusion, taking into account data obtained in this work, we can state that the firefly bioluminescent ATP assay allows obtaining the susceptibility to antibiotics for the most predominant bacteria isolated in Clinical Microbiology laboratories in only two hours. On the basis of this potential advantage, its future implementation in Clinical Microbiology laboratories could be taken into consideration.
Conflict of interestNone declared.
This work was supported by a grant (060/132421) from the Fundación Francisco Soria Melguizo, Spain.