In recent years it has been demonstrated that polyunsaturated fatty acids (PUFA) have anti-inflammatory and as regulators of lipid metabolism. However, the epigenomic mechanisms involved in these processes are not known in depth. The aim of this review was to describe the scientific evidence supports that regular consumption of PUFA may reduce obesity and overweight by altering epigenetic marks.
Material and methodsA search of recent publications was carried out in human clinical trials, as well as animal model and in vitro experiments.
ResultsExist a possible therapeutic effect of PUFAs on the prevention and development of obesity due to their ability to reversively modify the methylation of the promoters of genes associated with lipid metabolism and to modulate the activity of certain microRNAs.
ConclusionsA better knowledge and understanding of the PUFAs role in epigenetic regulation of obesity is possible with the current published results. The PUFAs may modulate the promotor epigenetic marks in several adipogenic genes and regulate the expression of several miRNAs.
En los últimos años se ha demostrado que los ácidos grasos poliinsaturados (AGPI) tienen efectos antiinflamatorios y como reguladores del metabolismo lipídico. No obstante, no se conocen en profundidad los mecanismos epigenómicos implicados en estos procesos. El objetivo de esta revisión fue describir las evidencias científicas que apoyan que el consumo regular de AGPI puede reducir la obesidad mediante modificaciones de las marcas epigenéticas.
Material y métodosSe realizó una búsqueda de publicaciones recientes llevadas a cabo en ensayos clínicos en humanos, modelos animales o ensayos in vitro.
ResultadosExiste un posible efecto terapéutico de los AGPI sobre la prevención y desarrollo de la obesidad gracias a su capacidad de modificar reversiblemente la metilación de los promotores de genes asociados con el metabolismo lipídico y de modular la actividad de determinados microARN.
ConclusionesLos resultados publicados hasta la fecha referentes al rol de los AGPI en la prevención de la obesidad contribuyen al mejor conocimiento y entendimiento de las modificaciones epigenéticas de la obesidad. Los AGPI han demostrado poder modificar epigenéticamente diferentes genes adipogénicos mediante la metilación de sus promotores o mediante la regulación de su interacción con diversos microARN.
Overweight and obesity are defined as an abnormal or excess fat accumulation that may impair health.1 This is a complex multifactorial disorder where both genetic and environmental factors interact. The body mass index (BMI) is the most commonly used tool for classifying overweight and obesity, and may be defined as the ratio between weight in kilograms and the square of height in meters (kg/m2).1 BMI values of 25 or higher represent overweight, while values of 30 or higher represent obesity.1 This measure correlates well with body adiposity. Excess weight is associated with increased morbidity and mortality, including an increased risk of type 2 diabetes mellitus, atherosclerosis, high blood pressure, hyperlipidemia, osteoarthritis, sleep apnea syndrome, and some types of cancer.2
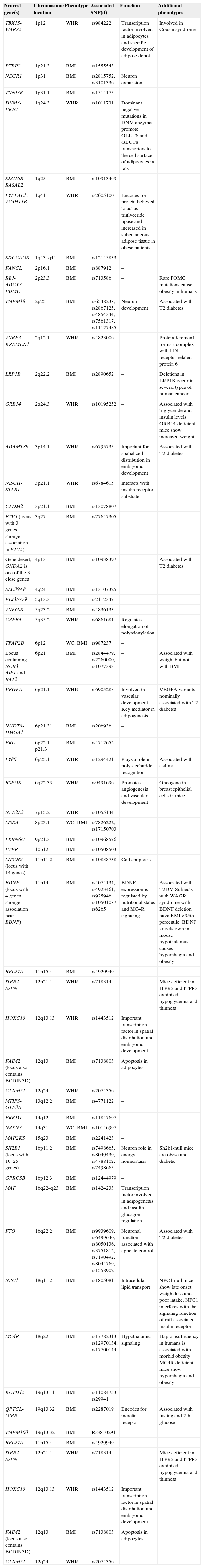
There is currently a pandemic of overweight and obesity which has been increasing for decades2 and continues to increase.3 A study of the worldwide population published in 2008 estimated that 23.2% of the adult population had overweight, and 9.8% obesity, which represents some 937 million and 396 million people with overweight and obesity respectively.3 This study also predicted for 2030 an adult population of up to 2160 million people with overweight and 1120 million with obesity if the secular trends seen to date continue.3 The situation is Spain is also worrying. A study published in 20114 reported a 34.2% prevalence of overweight in adults, with higher values in males (43.9%) as compared to females (25.7%). Obesity was reported in 13.6%, with no sex difference.4 This growing prevalence of obesity is related to an increased prevalence of metabolic syndrome.5 The definition of this syndrome, which is closely related to abdominal fat, usually refers to glucose intolerance, abdominal obesity, hypertension, and dyslipidemia that severely impair the health of those who suffer from it.5,6 Obesity is therefore a significant public health problem, and involves high financial costs because of its associated comorbidities.2 The worldwide financial burden of obesity has been estimated to range from 0.7% to 2.8% of all healthcare expenses, with a financial impact of 9.1% for overweight and obesity.2 The most commonly accepted model for explaining human obesity is based on the interaction between genetic predisposition, metabolic abnormalities, and environmental factors such as sedentary lifestyles and unhealthy nutrition. Specifically, it has been estimated from twin, adoption, and familial studies that the genetic component causes approximately 40% of interindividual variability in obesity values.7,8 More specifically, comparisons of twin studies with familial and adoption studies show that 60–90% of the BMI variance in the population may be explained by genetic effects.9 Linkage and association studies have located multiple obesity loci along the genome.10 The central role of lipid metabolism in obesity and overweight has led to extensive analysis of the genetic varieties of genes encoding for the proteins involved in the metabolic pathways of adipogenesis, energy intake, lipolysis, and energy expenditure. Thus, for example, polymorphisms in the apolipoprotein B11 and A5,11 CD36 (cluster of differentiation 36),12 USF1 (upstream transcription factor 1),13,14 FADS3 (fatty acid desaturase 3),14 GCKR (glucokinase regulatory protein),15 INSIG2 (insulin-induced gene 2),16 NPP1 (ectonucleotide pyrophosphatase/phosphodiesterase 1),17 FTO (fat mass and obesity-associated protein),18 and CTNNBL1 (catenin beta like 1)19 genes have been studied. More than 40 genetic variants associated with obesity and body fat distribution are currently known20 (Table 1). However, these studies with genetic markers cannot fully account for the heritability of obesity. This may partly be due to the polygenetic nature of obesity, in which different variants of the DNA sequence have only a small effect. For this reason, a very large analysis population is required for detection.10
The 54 loci associated with phenotypes of anthropometric obesity.
| Nearest gene(s) | Chromosome location | Phenotype | Associated SNP(d) | Function | Additional phenotypes |
|---|---|---|---|---|---|
| TBX15-WARS2 | 1p12 | WHR | rs984222 | Transcription factor involved in adipocytes and specific development of adipose depot | Involved in Cousin syndrome |
| PTBP2 | 1p21.3 | BMI | rs1555543 | – | |
| NEGR1 | 1p31 | BMI | rs2815752, rs3101336 | Neuron expansion | |
| TNNI3K | 1p31.1 | BMI | rs1514175 | – | |
| DNM3-PIGC | 1q24.3 | WHR | rs1011731 | Dominant negative mutations in DNM enzymes promote GLUT6 and GLUT8 transporters to the cell surface of adipocytes in rats | |
| SEC16B, RASAL2 | 1q25 | BMI | rs10913469 | – | |
| LYPLAL1; ZC3H11B | 1q41 | WHR | rs2605100 | Encodes for protein believed to act as triglyceride lipase and increased in subcutaneous adipose tissue in obese patients | |
| SDCCAG8 | 1q43–q44 | BMI | rs12145833 | – | |
| FANCL | 2p16.1 | BMI | rs887912 | – | |
| RBJ-ADCY3-POMC | 2p23.3 | BMI | rs713586 | – | Rare POMC mutations cause obesity in humans |
| TMEM18 | 2p25 | BMI | rs6548238, rs2867125, rs4854344, rs7561317, rs11127485 | Neuron development | Associated with T2 diabetes |
| ZNRF3-KREMEN1 | 2q12.1 | WHR | rs4823006 | – | Protein Kremen1 forms a complex with LDL receptor-related protein 6 |
| LRP1B | 2q22.2 | BMI | rs2890652 | – | Deletions in LRP1B occur in several types of human cancer |
| GRB14 | 2q24.3 | WHR | rs10195252 | – | Associated with triglyceride and insulin levels. GRB14-deficient mice show increased weight |
| ADAMTS9 | 3p14.1 | WHR | rs6795735 | Important for spatial cell distribution in embryonic development | Associated with T2 diabetes |
| NISCH-STAB1 | 3p21.1 | WHR | rs6784615 | Interacts with insulin receptor substrate | |
| CADM2 | 3p21.1 | BMI | rs13078807 | – | |
| ETV5 (locus with 3 genes, stronger association in ETV5) | 3q27 | BMI | rs77647305 | – | |
| Gene desert; GNDA2 is one of the 3 close genes | 4p13 | BMI | rs10938397 | – | Associated with T2 diabetes |
| SLC39A8 | 4q24 | BMI | rs13107325 | – | |
| FLJ35779 | 5q13.3 | BMI | rs2112347 | – | |
| ZNF608 | 5q23.2 | BMI | rs4836133 | – | |
| CPEB4 | 5q35.2 | WHR | rs6861681 | Regulates elongation of polyadenylation | |
| TFAP2B | 6p12 | WC, BMI | rs987237 | – | |
| Locus containing NCR3, AIF1 and BAT2 | 6p21 | BMI | rs2844479, rs2260000, rs1077393 | – | Associated with weight but not with BMI |
| VEGFA | 6p21.1 | WHR | rs6905288 | Involved in vascular development. Key mediator in adipogenesis | VEGFA variants nominally associated with T2 diabetes |
| NUDT3-HMGA1 | 6p21.31 | BMI | rs206936 | – | |
| PRL | 6p22.1–p21.3 | BMI | rs4712652 | – | |
| LY86 | 6p25.1 | WHR | rs1294421 | Plays a role in polysaccharide recognition | Associated with asthma |
| RSPOS | 6q22.33 | WHR | rs9491696 | Promotes angiogenesis and vascular development | Oncogene in breast epithelial cells in mice |
| NFE2L3 | 7p15.2 | WHR | rs1055144 | – | |
| MSRA | 8p23.1 | WC, BMI | rs7826222, rs17150703 | – | |
| LRRN6C | 9p21.3 | BMI | rs10968576 | – | |
| PTER | 10p12 | BMI | rs10508503 | – | |
| MTCH2 (locus with 14 genes) | 11p11.2 | BMI | rs10838738 | Cell apoptosis | |
| BDNF (locus with 4 genes, stronger association near BDNF) | 11p14 | BMI | rs4074134, rs4923461, rs925946, rs10501087, rs6265 | BDNF expression is regulated by nutritional status and MC4R signaling | Associated with T2DM Subjects with WAGR syndrome with BDNF deletion have BMI >95th percentile. BDNF knockdown in mouse hypothalamus causes hyperphagia and obesity |
| RPL27A | 11p15.4 | BMI | rs4929949 | – | |
| ITPR2-SSPN | 12p21.1 | WHR | rs718314 | – | Mice deficient in ITPR2 and ITPR3 exhibited hypoglycemia and thinness |
| HOXC13 | 12q13.13 | WHR | rs1443512 | Important transcription factor in spatial distribution and embryonic development | |
| FAIM2 (locus also contains BCDIN3D) | 12q13 | BMI | rs7138803 | Apoptosis in adipocytes | |
| C12orf51 | 12q24 | WHR | rs2074356 | – | |
| MTIF3-GTF3A | 13q12.2 | BMI | rs4771122 | – | |
| PRKD1 | 14q12 | BMI | rs11847697 | – | |
| NRXN3 | 14q31 | WC, BMI | rs10146997 | – | |
| MAP2K5 | 15q23 | BMI | rs2241423 | – | |
| SH2B1 (locus with 19–25 genes) | 16p11.2 | BMI | rs7498665, rs8049439, rs4788102, rs7498665 | Neuron role in energy homeostasis | Sh2b1-null mice are obese and diabetic |
| GPRC5B | 16p12.3 | BMI | rs12444979 | – | |
| MAF | 16q22–q23 | BMI | rs1424233 | Transcription factor involved in adipogenesis and insulin-glucagon regulation | |
| FTO | 16q22.2 | BMI | rs9939609, rs6499640, rs8050136, rs3751812, rs7190492, rs8044769, rs1558902 | Neuronal function associated with appetite control | Associated with T2 diabetes |
| NPC1 | 18q11.2 | BMI | rs1805081 | Intracellular lipid transport | NPC1-null mice show late onset weight loss and poor intake. NPC1 interferes with the signaling function of raft-associated insulin receptor |
| MC4R | 18q22 | BMI | rs17782313, rs12970134, rs17700144 | Hypothalamic signaling | Haploinsufficiency in humans is associated with morbid obesity. MC4R-deficient mice show hyperphagia and obesity |
| KCTD15 | 19q13.11 | BMI | rs11084753, rs29941 | – | |
| QPTCL-GIPR | 19q13.32 | BMI | rs2287019 | Encodes for incretin receptor | Associated with fasting and 2-h glucose |
| TMEM160 | 19q13.32 | BMI | Rs3810291 | – | |
| RPL27A | 11p15.4 | BMI | rs4929949 | – | |
| ITPR2-SSPN | 12p21.1 | WHR | rs718314 | – | Mice deficient in ITPR2 and ITPR3 exhibited hypoglycemia and thinness |
| HOXC13 | 12q13.13 | WHR | rs1443512 | Important transcription factor in spatial distribution and embryonic development | |
| FAIM2 (locus also contains BCDIN3D) | 12q13 | BMI | rs7138803 | Apoptosis in adipocytes | |
| C12orf51 | 12q24 | WHR | rs2074356 | – |
WC: waist circumference; BMI: body mass index; POMC: WHR: waist/hip ratio.
Another potential explanation is the existence of other forms of variation, such as epigenetic modifications and alterations.20 Epigenetics may currently be defined as the heredity of DNA activity that does not depend on the sequence itself, but on chemical modifications in DNA and adjacent regulatory proteins.21 The best known epigenetic marks include the addition of a methyl group to DNA in cytosine of the CpG dinucleotide.21 These dinucleotides are abundant in the promoter regions of many genes. Hypermethylation is usually associated with decreased gene expression (silencing); by contrast, hypomethylation is associated with increased expression.22,23 The concept of genetic imprinting is related to the DNA methylation level. This concept describes the heredity of specific epigenetic information from one of the parents. Some genes acquire a maternal or paternal imprint during gametogenesis and, as a result, are widely expressed from a single allele during embryonal development and in adult tissues.24 Defective genetic imprinting is associated with developmental disorders and clinical phenotypes, among which abnormal body weight is usually included.24 A well known example is Prader–Willi syndrome, characterized by cognitive impairment and voracious and uncontrollable appetite, which is often associated with the development of severe obesity in the first six years of life.24 An additional epigenetic mark studied is the modification of the proteins called histones. In addition to packaging DNA, histones play a very significant role in post-translational modifications of their amino acids (e.g. lysine acetylation, arginine methylation, serine phosphorylaton)21 Other epigenetic marks under study are defined by the arrangement of high-order structures formed by DNA-histone complexes (the so-called nucleosomes) and the activity of non-coding RNAs such as microRNAs, interference RNAs, long-chain non-coding RNAs, and antisense RNAs, amongst others.25,26 These non-coding RNAs regulate post-transcriptionally gene expression through their pairing to the 3′ untranslated region (3′ UTR) of messenger RNA.27 For example, miR-33 and miR-122 control triglyceride metabolism and cholesterol biosynthesis in mouse liver, and suggest that their dysregulation is directly associated with the development of metabolic diseases such as obesity and metabolic syndrome.28,29 The implication of long-chain non-coding RNAs (lncRNAs) in adipose tissue plasticity and the regulation of adipogenesis is also known.30,31
As previously reported, obesity is a multifactorial, polygenic disorder where genetic and epigenetic factors interact with environmental factors such as physical activity, alcohol, and smoking. However, nutrition is probably the most important factor.32 In addition, epigenetic changes show a great plasticity and respond to environmental signals, including diet.33 Because of the influence of maternal metabolism on embryo development during pregnancy, it has been suggested that the nutritional status of the mother during pregnancy may induce epigenetic dysfunctions in the newborn.34–40 Although epigenoma involvement occurs at specific time periods, in the first stages of embryogenesis and infancy, intervention in adult age is also possible.33 Exposure to diets rich or deficient in given nutrients for long time periods (years) has been seen to induce epigenetic changes with consequences for health and the risk of disease.33 Thus, polyphenols exert their antilipidemic and antiatherogenic activity not only by regulating the expression of different genes associated with the immune system and energy metabolism, but also by inducing changes in the methylation pattern of CpG islands of DNA,41,42 histone acetylation,43 and the modulation of expression of some miRNAs44 in adults. In this regard, Joven et al.45 used hyperlipidemic mice with LDL receptor deficiency to assess the role of polyphenols in the prevention of metabolic disease through the regulation of expression of the hepatic microRNAs miR-103/107 and miR-122. In their results, they stressed that oral polyphenol administration reversed the changes caused in the non-specific microRNAs miR-103/107 after chronic polyphenol intake, along with the lack of response of the specific miRNA miR-122, and speculated about a potential implication of polyphenols in cell metabolism in the liver. They also postulated that the modulation of microRNA expression could be a significant additional mechanism of intervention in chronic diseases. Despite the foregoing, additional studies are required in humans to elucidate the epigenetic effects of polyphenols and other components such as long-chain PUFAs.
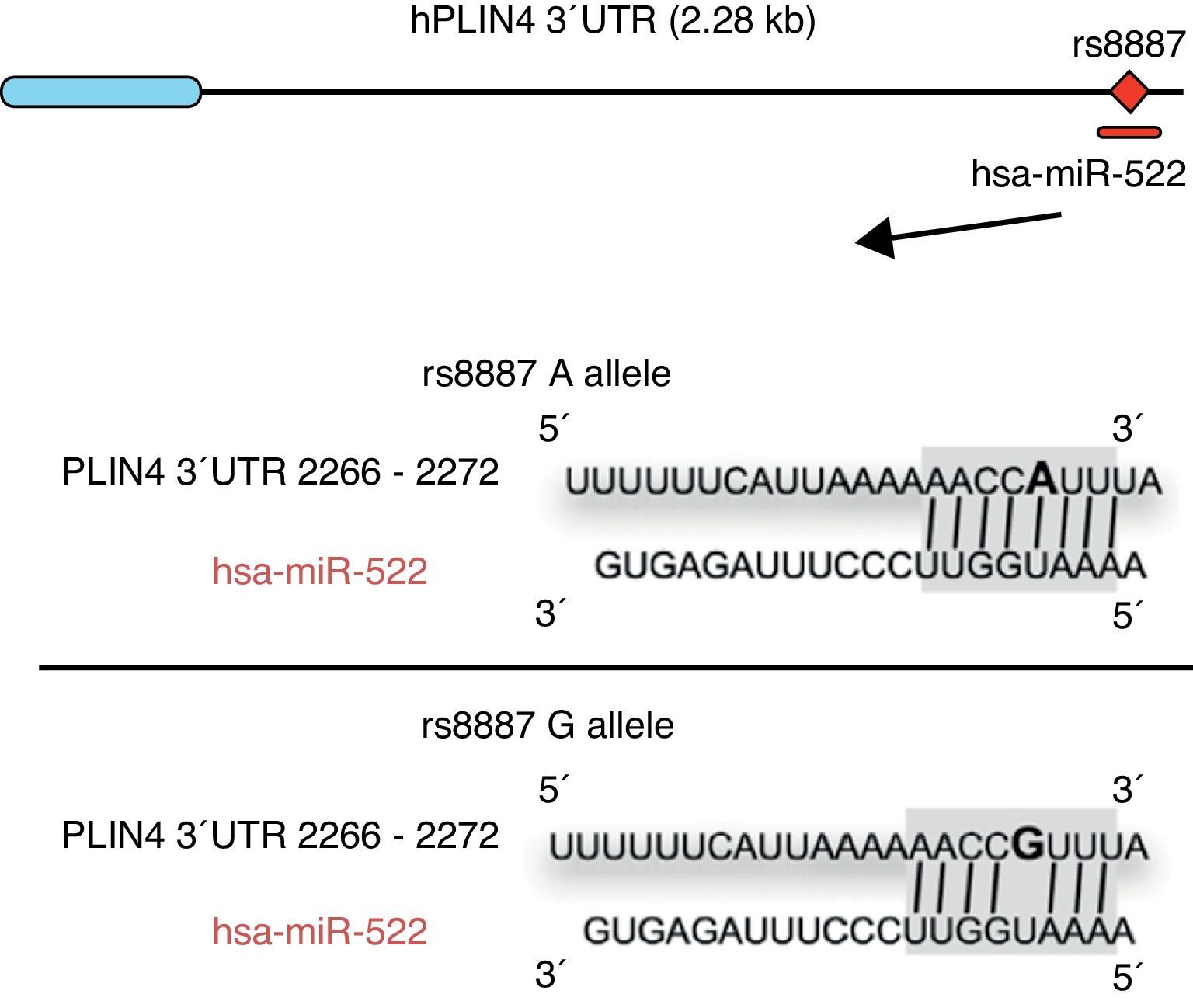
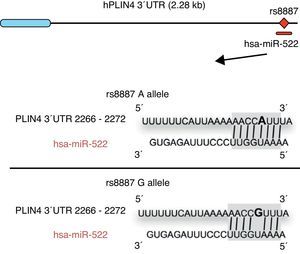
Omega-3 fatty acids (n-3) have been related to various properties and therapeutic uses in humans.46–53 Thus, intake of the recommended amounts of n-3 compounds docosahexaenoic (DHA) and eicosapentaenoic acids decreases the risk of death and coronary diseases by preventing arrhythmia, the formation of prostaglandins and leukotriene precursors, the inhibition of inflammatory cytokines, the promotion of lipolysis, and fatty acid oxidation, as well as the inhibition of lipogenesis and a reduction in total triglycerides and very low density lipoproteins (VDVDLc). Diets with high n-3 contents have recently been seen to decrease the risk of the development of different types of cancer (e.g. colorectal and breast cancer) and their cell proliferation, among other properties.54–58 The molecular processes associated with antilipidemic and antiatherogenic properties, as well as the anti-inflammatory and anti-cell development, of n-3 fatty acids result from their ability to regulate the expression of different genes associated with the immune system and energy metabolism,59–62 or their epigenetic regulation capacity through the induction of changes in the methylation pattern of CpG islands of DNA,63 and the modulation of expression of some miRNAs.64–66 In this regard, for example, it has been reported that n-3 PUFAs modify the interaction between miR-522 and the 3′ UTR region of the perilipin 4 gene (PLIN4), resulting in a change in obesity-related phenotypes (Fig. 1).67 However, few studies are available reporting the effect of the intake of different types of polyunsaturated fatty acids (PUFAs) on epigenetic modification and the resulting genetic expression. Consequently, there is a need to verify the public data and to illustrate the relationship between the intake of PUFAs, especially n-3, and epigenetic modifications. The purpose of this study was therefore to review the most recent studies on the effects of PUFA intake and the risk of obesity or overweight, in an attempt to elucidate the associated epigenetic mechanisms, especially DNA methylation and the role of non-coding RNAs.
Minor A allele rs8887 creates a new miR-522 MRE in the PLIN4 3¿ UTR gene. miR-522 diagram: PLIN4 3¿ UTR sequences with the A or G allele. The miR-522 site is shaded gray, and variant rs8887 appears in bold.
In this review, a search of recent publications was made in the following specialized electronic databases: NCBI, Elsevier Journal, Scielo, Science Direct, Springer Link. The results from studies conducted in vitro, in animal models, and in humans were collected. Reviews collecting and analyzing the effectiveness of PUFAs in certain treatments, such as antihypertensive and lipid lowering therapies, among others, were also included. Epigenetic concepts related to non-coding RNAs and chemical modifications in histones, obesity, high blood pressure, and atherosclerosis were also analyzed to describe in greater detail the potential epigenetic mechanisms of PUFAs. The following keywords were used: polyunsaturated fatty acids, histone acetylaton, DNA methylation, microRNA, epigenetics, obesity, overweight, and metabolic syndrome. A total of 84 articles were revised, including reviews. The articles selected were divided into the following categories: (1) generic articles on epigenetics, obesity, and PUFAs; (2) articles on the relationship between PUFA consumption and DNA methylation, histone acetylation, and non-coding RNA modulation.
Results and discussionFew reports are available on the epigenetic effect of the intake of n-3 and n-6 PUFAs and their role in obesity control and prevention. Studies analyzing the effects of PUFAs on epigenetic modifications used in this review were grouped based on the type of epigenetic mark: (1) the addition of a methyl group to DNA at the cytosine in the CpG dinucleotide; (2) modification of the so-called histone proteins; (3) modification of non-coding RNA expression. The most relevant results and conclusions are specified for each of them.
As stated above, different demethylation waves occur during the first few days of embryo development, followed by increased de novo methylation in embryo and extraembryonic tissues such as the placenta.68,69 Guo et al.68 showed that the greatest demethylation wave is completed in the two-cell stage. Soon after this implantation, a remethylation wave occurs, and epigenetic patterns are established for the different cell types.33,68 During pregnancy, there may be a first contact between the embryo and nutrients or secondary metabolites from the mother, influencing fetal epigenoma and increasing or decreasing the risk of developing some diseases. Kulkarni et al.63 reported that supplementation with n-3 (45g of fish oil and 25g of soybean oil per kg of diet) to pregnant rats combined with excess folic and vitamin B12 deficiency increased DNA methylation in the placenta to control levels. Thus, decreased DNA methylation levels in rat placenta were reversed when the diet was supplemented with DHA, showing that DHA levels play a very important role in determining placental methylation levels. These results are consistent with those obtained in studies in animal models where n-3 supplementation during pregnancy70 or during the first days after birth71 was able to prevent or reduce the adverse effects of fetal programming. In obese adult mice, Fan et al.72 showed that the regulation of expression of leptin, leptin receptor, and the neuropeptide precursor proopiomelanocortin (POMC) genes was modified by diet supplementation with n-3 (35g/kg of soybean oil; 17.5g/kg of soybean oil and 17.5g/kg of fish oil; 35g/kg of fish oil for each of the three groups with no n-3 deficiency), but that the methylation of the promoters of those genes did not change.72 Other studies conducted in animal models have shown that the effect of n-3 supplementation on DNA methylation depends on the gene and tissue studied, particularly during pregnancy and lactation.63,73,74 Thus, Niculescu et al.74 were able to show an association between the availability of α-linolenic acid (ALA; supplementation of 75,367nmol/mg/day) during pregnancy and lactation in mice and changes in DNA methylation of the FADS2 gene (fatty acid desaturase 2) and intron number 1 in livers from dams and pups at the end of the lactation period. FADS2 is a desaturase enzyme that catalyzes the different steps in the biosynthetic pathway of long-chain PUFAs from linoleic acid (n-6) and ALA.75 Moreover, this study suggested that maternal interaction with ALA during pregnancy and lactation could differentially alter n-3 and n-6 metabolism.74 In humans, a recent study conducted in young women with overweight treated with a calorie restricted diet showed that supplementation with n-3 derived from fish oil (>1300mg/day as 6 capsules daily) induced small epigenetic changes that decreased DNA methylation of the CD36 gene of mononuclear cells in blood after adjustment for the body weight of the women.76
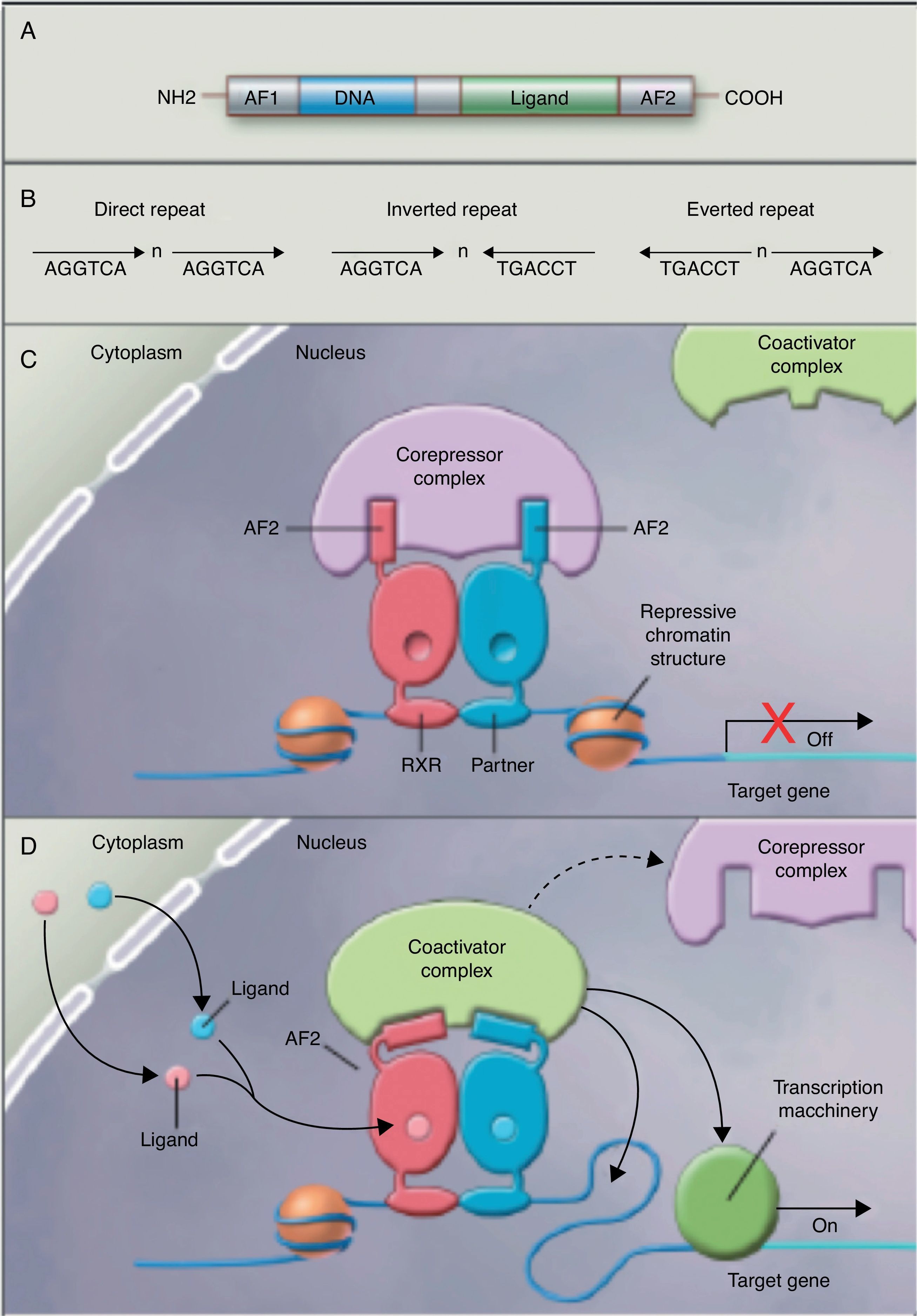
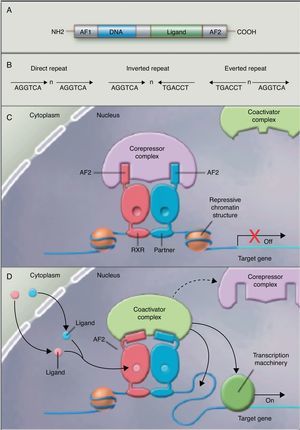
No studies were found relating the effects of the intake of PUFAs and their metabolites to histone acetylation and the resultant chromatin remodeling, which is important for the expression of genes of the nuclear receptor superfamily associated with the control and development of obesity and lipid metabolism.77 Although no studies are available on the subject, some authors78 are coming round to the idea that long-chain PUFAs could make it possible to control the expression of PPAR (peroxisome proliferator-activated receptor gamma) and its target genes through sequential chromatin remodeling. In other words, PUFA intake could modify the multiprotein corepressor complex with histone deacetylase activity, modify chromatin remodeling, permitting transcription factor binding to its promoter, facilitating its transcription and the expression of all target genes, many of which are related to obesity (Fig. 2).
Nuclear receptors as ligand-dependent transcription factors. (A) Canonical structure of a nuclear response element (NRE) including the n N-terminal activation function (AF1), DNA binding, ligand binding, and C-terminal domains (AF2). (B) Number of nucleotides between the central elements (n) that confer additional specificity. (C, D) Heterodimer without and with ligand associated with the corepressor and coactivator complex.
Finally, we report on studies showing the regulation of non-coding RNAs by PUFAs and its potential implications in obesity. The first example of a genetic variant that results in a binding site for a microRNA (miRNA) which influences the traits related to obesity through a gene–diet interaction modulated by n-3 PUFAs was recently shown.67 miRNAs are small non-coding transcripts consisting of approximately 21–25 nucleotides. They play a determinant role in the regulation of genes associated with processes of cell differentiation and development, the proliferation and maintenance of homeostasis, amongst others. These miRNAs, associated with multienzyme complexes, are guided for the recognition of complementary sequences in the 3′ UTR or 5′ UTR region of mRNA.79 Their interaction usually leads to mRNA degradation and translational repression, with a subsequent reduction in protein activity. Richardson et al.67 investigated the relationship between 7 SNPs in the PLIN4 gene (rs8887, rs11673616, rs892158, rs7250947, rs8102428, rs1609717, rs884164) and obesity-related phenotypes from samples of subjects from two populations of European ancestry.67 These authors conducted a meta-analysis which showed significant interactions between the rs8887 polymorphism for the minor A allele of the PLIN4 gene, the intake of n-3 PUFAs, and anthropometric measurements. PLIN4 is a protein of the PAT family with a great affinity for lipid storage80 droplets, which have an influence on the risk of developing metabolic diseases.81 The authors also reported that, at the structural level, the presence of the A allele in the 3′ untranslated region (3′ UTR) of the PLIN4 gene created a molecular recognition element (MRE) for miR-522, which did not occur in the case of the G allele (Fig. 1). Data provided by this study show that high n-3 intake may induce in allele A carriers decreased anthropometric values as compared to non-carriers, and specifically to homozygotes for the G allele, because there is no interaction in them between miRNA and the 3′ UTR region of the PLIN4 gene.67 Decreased PLIN4 gene expression due to miR-522 may contribute to obesity-related phenotypes, but additional studies are required to confirm this, and to ascertain whether the proposed mechanism may be operative for other miRNAs. In another recent study, Baselga et al.82 were able to counteract the dyslipidemic effect of two miRNAs by supplementing the diet of obese rats with proanthocyanidins and DHA. The miRNAs analyzed (miR-122 and miR-33a) are important regulators of lipid metabolism in the liver.83 The study objective was to assess whether liver levels of miR-122 and miR-33a correlated with lipidemia induced by nutrition in different rat models. To do this, liver levels of both miRNAs were measured in dyslipidemic rats fed a cafeteria diet (CD) with no supplementation and rats fed a CD supplemented with proanthocyanidins or DHA. The CD was shown to increase miR-122 and miR-33a levels in the liver. By contrast, levels of both miRNAs were reversed in rats with DHA supplementation, with an even greater reduction in rats supplemented with both compounds (proanthocyanidins and DHA). With regard to the lipid profile, long-term treatment with proanthocyanidins improved the atherogenic index altered by CD, normalizing plasma triglyceride (TG) and LDL levels, and also decreased total lipid and TG levels in the liver. By contrast, rats fed CD supplemented with DHA showed a normalization of plasma total cholesterol and LDL levels, but the lipid content in the liver was not affected. The concomitant administration of both treatments (polyphenols and DHA) had a lipid-lowering effect, with decreases in liver and plasma levels similar to those achieved by individual treatments alone. The authors concluded that their effect was complementary, rather than synergistic or additive, but further studies are needed to elucidate the mechanism by which proanthocyanidins and DHA repress miR-122 and miR-33a.82
ConclusionsPUFA intake has been associated with different therapeutic properties. Specifically, based on the results of this review, we conclude that PUFA intake may control the parameters related to obesity through different epigenetic mechanisms. The early results suggest that PUFAs are able to reversibly modify the methylation of adipogenic gene promoters and, thus, their expression. This is a remarkable result, because epigenetic changes may be one of the weak points in the development of obesity, as we can inactivate or activate epigenetically inactivated genes using adequate nutrients. There is currently no information on genetic modulation through alternative epigenetic mechanisms, such as histone modifications. However, according to the early results, the potential association between PUFAs and the repression of the expression of genes associated with lipid metabolism by miRNAs is starting to become evident in animal models.
The results published to date do not allow us to determine a dose of PUFAs in terms of its therapeutic properties. However, the results do represent interesting findings which should be thoroughly analyzed, because understanding of the distribution and function of PUFAs in obese patients may be helpful in achieving effective treatment. Continued research in the field of alternative non-drug treatments, such as functional foods, is also required. Only a limited number of PUFAs have been tested to date, and since the effects of the different components are not equivalent, the results cannot be generalized. Future large scale studies with control of doses, active components, bioavailability, and other critical variables, such as genetic background, will therefore be crucial for providing the scientific evidence required to ascertain the epigenetic modifications induced by PUFAs and their contribution to obesity development and prevention.
Conflicts of interestThe authors state that they have no conflicts of interest.
Please cite this article as: Hernando Boigues JF, Mach N. Efecto de los ácidos grasos poliinsaturados en la prevención de la obesidad a través de modificaciones epigenéticas. Endocrinol Nutr. 2015;62:338–349.