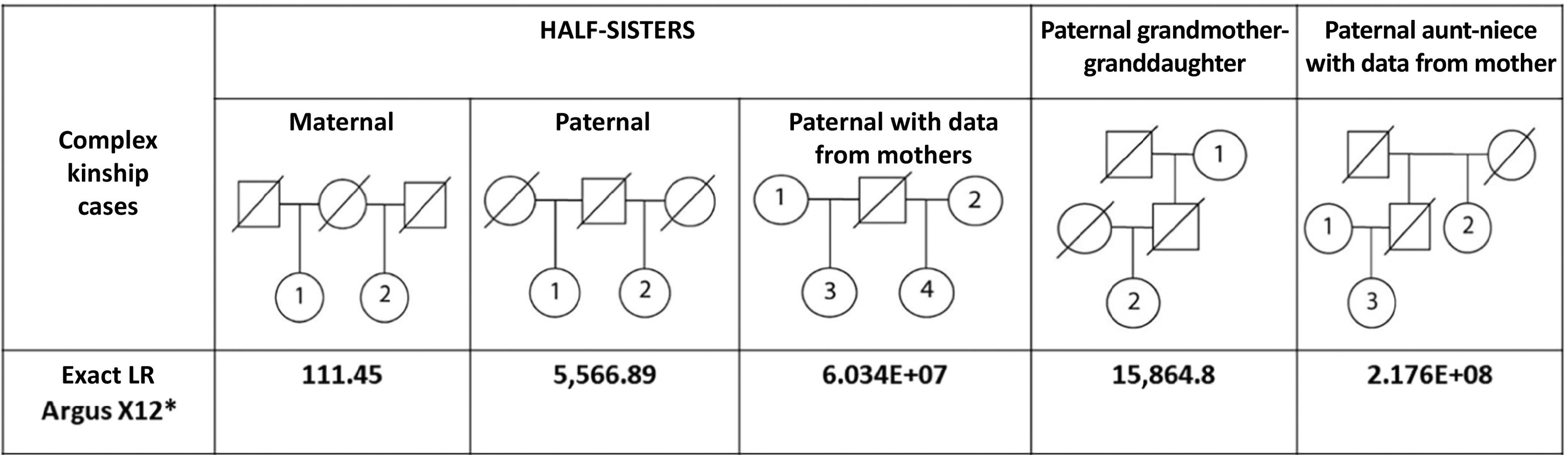
The Investigator Argus X-12 kit is a valuable complementary tool for human identification purposes, especially for solving complex kinship cases. The analysis of these cases generates valuable information in routine forensic work, such as a posteriori informativeness (LR: likelihood ratio) and mutation rates.
AimTo analyze the LR offered by the Argus X-12 QS kit in 74 Mexican families, including the father, mother, and daughter(s), respectively.
MethodsExact LRs were estimated with the FamlinkX software in the families as paternity trio cases (n = 74; average 8.E+14) and duo cases (n = 148; average 6.1E+11), omitting the mother and father, respectively.
ResultsThe LR of the trios (a posteriori) was significantly higher than the -a priori- parameter IP typical (3.98E+04). As a reference, we report the LR of complex kinship cases solved with X-STRs: maternal half-sisters (LR = 111), paternal half-sisters (LR = 5567), paternal half-sisters with their mothers (LR = 6.0E+07, and paternal grandmother-granddaughter (LR = 15,864). Nine mutations were detected in 161 meiosis. The mutation rate was estimated in seven X-STRs, which was 0.0062 (1/161) for DXS10148, DXS10135, DXS8378, DXS10146, and DXS7423, and 0,0124 (2/161) for DXS10074 and DXS10101. These findings will be useful in future meta-analyses to achieve more conclusive estimates. A null allele in DXS10148 was inferred between a mother and daughter, supported by the low probability that the finding is explained by an 8-step mutation (26.1 → 18), and the low frequency of the 26.1 homozygote.
ConclusionThe a posteriori informativeness of Argus X-12 in paternity and kinship cases described herein justifies the inclusion of the X-STRs in forensic genetics labs.
El kit Investigator Argus X-12 es una herramienta complementaria valiosa con fines de identificación humana, especialmente para resolver casos de parentesco complejos. El análisis de éstos casos de parentesco genera información valiosa en el trabajo de rutina forense, como informatividad a posteriori (LR: likelihood ratio) y las tasas de mutación.
ObjetivoAnalizar el LR que ofrece el kit Argus X-12 QS en 74 familias mexicanas, incluyendo al padre, madre e hija(s).
MétodosSe estimaron LR exactos con el software FamlinkX en las familias como casos de paternidad tipo trío (n = 74; promedio 8.E+14) y tipo dúo (n = 148; promedio 6.1E+11), al no incluir a la madre y al padre, respectivamente.
ResultadosEl LR de los tríos (a posteriori) resultó ser significavamente mayor que el parámetro -a priori- IP típico (3.98E+04). Como referencia, se reporta el LR de casos de parentesco complejo resueltos con X-STRs: media hermandad via materna (LR = 111), media hermandad paterna (LR = 5567), medias hermanas paternas y sus madres (LR = 6.0E+07) y abuela-paterna-nieta (LR = 15,864). Se detectaron 9 mutaciones en 161 meiosis, con las que se estimó la tasa de mutación en siete X-STRs, en el rango de 0,0062 (1/161) para DXS10148, DXS10135, DXS8378, DXS10146 y DXS7423, hasta 0,0124 (2/161) para DXS10074 y DXS10101. Estos hallazgos serán de utilidad en futuros metaanálisis para lograr estimaciones más concluyentes. Se infirió un alelo nulo en DXS10148 entre una madre e hija, sustentado por la baja probabilidad de una mutación materna de 8 pasos (26.1 → 18; p = 9.15E−05), y la baja frecuencia poblacional del homocigoto 26.1.
ConclusiónLa informatividad a posteriori del kit Argus X-12 en casos de paternidad reales y los casos de parenteso descritos, justifican la inclusión de los X-STRs en los laboratorios de genética forense.
In recent decades, X-chromosome Short Tandem Repeats (X-STR) markers have been integrated into the battery of markers available in forensic genetics laboratories for human identification purposes.1 This is due to their characteristic inheritance pattern, which is useful for solving complex kinship cases in which at least one woman is involved, such as the paternal grandmother-granddaughter relationship without the father, paternal brotherhood or half-sisterhood between women, ruling out incest cases, and in various scenarios during the identification of missing persons with a small number of available relatives.2,3 The biostatistical interpretation of kinship relationships with X-STR requires the use of allele and haplotype frequencies, in addition to considering genetic linkage and linkage disequilibrium (LD) between pairs of loci,4,5 which are taken into account by the FamLinkX software for the estimation of kinship indices or Likelihood Ratios (LRs).6 Furthermore, the mutation rate per X-STR is necessary to explain isolated exclusions in kinship assessment and to avoid false conclusions.7,8
The Investigator Argus X-12 system (Qiagen) analyses 12 X-STRs grouped into 4 linkage groups with 3 X-STRs each.1,3 This is probably the most widely used X-STR-based kit, as many population data required for forensic interpretation are available with the FamlinkX software (http://www.famlink.se/fx_download.html), which has facilitated its implementation in any laboratory. Most population studies in forensic genetics report different a priori parameters, such as heterozygosity (Het), polymorphism information content (PIC), typical paternity index (PI), discrimination power (PD) and mean exclusion probability (MEC), in addition to allele and haplotype frequencies.9,10 However, a posteriori parameters of real paternity cases -such as LR- are scarcely reported worldwide, and they could be useful to indicate the real contribution of genetic systems to forensic routine work.11,12 In this study, based on Mexican families with confirmed kinship including father, mother, and daughters, the a posteriori informativeness offered by the Argus X-12 system was estimated through the exact LR estimated with FamlinkX. These results are discussed in the context of the LR obtained from 5 cases of complex kinship between women involving different types of half-siblingships, the paternal grandmother-granddaughter relationship and between a paternal aunt-niece in the presence of the mother. Finally, among the findings within these Mexican families, a null allele and 9 exclusions involving mutations are described, which allowed estimating mutation rates in the 7 X-STRs involved. Although the sample analyzed is clearly insufficient to estimate mutation rates, these findings are useful in future meta-analyses to achieve reliable estimates.
MethodologySampleBlood samples were collected on FTA paper from 74 families including father, mother, and daughters, giving a total of 235 samples (161 meiosis) (Supplementary Material S1). In addition to the analysis with the Argus X-12 kit, their relationship was previously confirmed with 22 autosomal STRs included in the PowerPlex® Fusion system (Promega Corp.). Small 1.2 mm punches of the blood spot on paper were washed with FTA Purification Reagent (Whatman™) and DNAse-free water, which were used directly as template DNA for subsequent analyses.
X-STR genotypePCR amplification was carried out with the Investigator Argus X-12 QS kit (Qiagen, Hilden, Germany), according to the instructions described by the manufacturer. The amplification products were subjected to capillary electrophoresis on an ABI-Prism 3130 genetic analyzer (Applied Biosystems®, Foster City, USA). Genotype assignment was performed using Genemapper software version 3.2 (Applied Biosystems®) with the aid of the allelic ladder included in the Argus X-12 QS kit. Our laboratory participates in the quality control exercise organized annually by the Spanish and Portuguese Speaking Group of the International Society for Forensic Genetics (www.ghep-isfg.org), including the results of the Argus X-12 system.
Data analysisFor each of the 74 Mexican families, the exact LRs were estimated by X-STR marker, by linkage group, and the combined LR in the 74 standard trio cases, as well as in 148 cases configured as duos by not including the father and then omitting the mother (Supplementary Material S2). It should be noted that estimating an LR twice in the same daughter in duo cases does not duplicate these values, since the LR via the paternal route involves different alleles and haplotypes than the LR via the maternal route. The FamlinkX software was used to calculate the LRs,6 using the published Mexican population database and the default lambda value (λ = 1000).9 Using the same FamlinkX database,9 the LRs of 5 complex paternity cases were estimated in which X-STRs are used: (1) maternal half-siblingship between 2 women; (2) paternal half-siblingship between 2 women; (3) paternal half-siblingship in the presence of the mothers; (4) paternal grandmother-granddaughter; and (5) paternal aunt-niece in the presence of the mother. The complete genotype of these 5 cases for the Argus X-12 kit is presented in Supplementary Material S3.
Finally, based on the isolated non-concordances observed in some of the Mexican families (Supplementary Material S1), the mutation rate was estimated considering the total number of meioses for the loci involved. In addition, it was possible to describe in most cases the parental origin of the mutation (maternal or paternal), the number of mutation steps, and whether the mutation involved gain or loss.
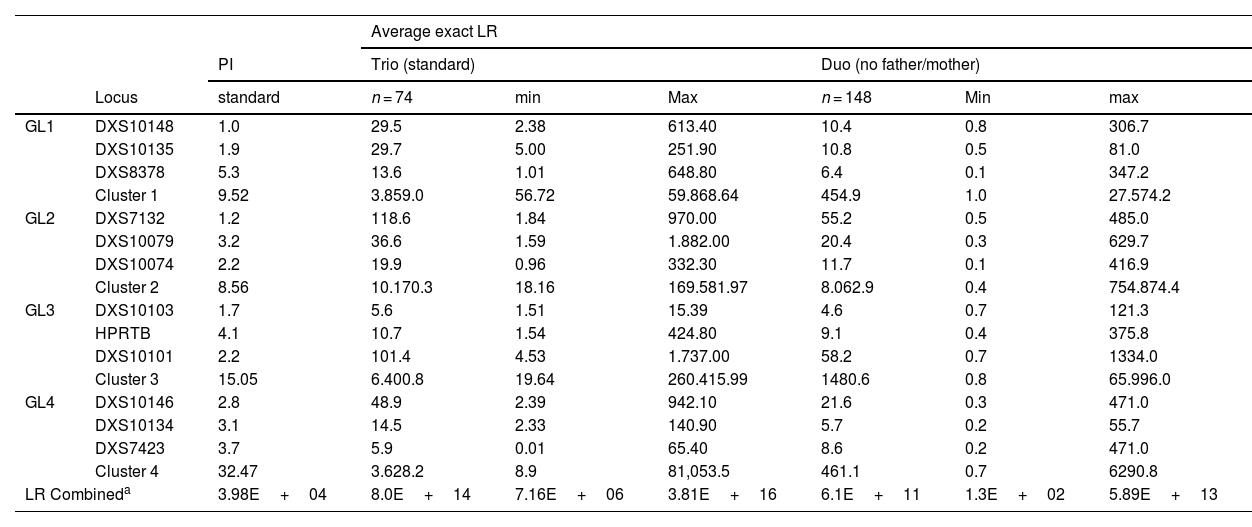
Results and discussionsInformativeness of a posteriori kinship (LR)Generally, population studies in forensic genetics evaluate the informativeness of a locus and/or genetic system using a priori forensic parameters or simulations. However, the analysis of informativeness values of cases solved a posteriori (LR) also provides valuable data, even from a single family.10–12 In this study, in 74 Mexican families with proven kinship, we estimated different a posteriori kinship parameters with FamlinkX based on the Argus X-12 kit, configured as 74 trio paternities and 148 duo paternities (Table 1). To our knowledge, this is the first report in which the a posteriori informativeness offered by the Argus X-12 kit is evaluated in Latin America.
Typical paternity indices (PIs) and exact likelihood ratios (LRs) obtained in 74 Mexican families organized as trios and duos, with the FamLinkX software from the 12 X-STRs included in the Argus X-12 QS system and linkage groups (LGs).
| Average exact LR | ||||||||
|---|---|---|---|---|---|---|---|---|
| PI | Trio (standard) | Duo (no father/mother) | ||||||
| Locus | standard | n = 74 | min | Max | n = 148 | Min | max | |
| GL1 | DXS10148 | 1.0 | 29.5 | 2.38 | 613.40 | 10.4 | 0.8 | 306.7 |
| DXS10135 | 1.9 | 29.7 | 5.00 | 251.90 | 10.8 | 0.5 | 81.0 | |
| DXS8378 | 5.3 | 13.6 | 1.01 | 648.80 | 6.4 | 0.1 | 347.2 | |
| Cluster 1 | 9.52 | 3.859.0 | 56.72 | 59.868.64 | 454.9 | 1.0 | 27.574.2 | |
| GL2 | DXS7132 | 1.2 | 118.6 | 1.84 | 970.00 | 55.2 | 0.5 | 485.0 |
| DXS10079 | 3.2 | 36.6 | 1.59 | 1.882.00 | 20.4 | 0.3 | 629.7 | |
| DXS10074 | 2.2 | 19.9 | 0.96 | 332.30 | 11.7 | 0.1 | 416.9 | |
| Cluster 2 | 8.56 | 10.170.3 | 18.16 | 169.581.97 | 8.062.9 | 0.4 | 754.874.4 | |
| GL3 | DXS10103 | 1.7 | 5.6 | 1.51 | 15.39 | 4.6 | 0.7 | 121.3 |
| HPRTB | 4.1 | 10.7 | 1.54 | 424.80 | 9.1 | 0.4 | 375.8 | |
| DXS10101 | 2.2 | 101.4 | 4.53 | 1.737.00 | 58.2 | 0.7 | 1334.0 | |
| Cluster 3 | 15.05 | 6.400.8 | 19.64 | 260.415.99 | 1480.6 | 0.8 | 65.996.0 | |
| GL4 | DXS10146 | 2.8 | 48.9 | 2.39 | 942.10 | 21.6 | 0.3 | 471.0 |
| DXS10134 | 3.1 | 14.5 | 2.33 | 140.90 | 5.7 | 0.2 | 55.7 | |
| DXS7423 | 3.7 | 5.9 | 0.01 | 65.40 | 8.6 | 0.2 | 471.0 | |
| Cluster 4 | 32.47 | 3.628.2 | 8.9 | 81,053.5 | 461.1 | 0.7 | 6290.8 | |
| LR Combineda | 3.98E+04 | 8.0E+14 | 7.16E+06 | 3.81E+16 | 6.1E+11 | 1.3E+02 | 5.89E+13 | |
The X-STRs DXS7132 and DXS10101 consistently obtained the highest LR values, both in trio-type and duo-type paternities (Table 1). In accordance with the higher informativeness of these X-STRs, their linkage groups GL2 and GL3 generated the highest LR values, respectively. Regarding the estimated LR in the X-STRs, in the trios, this value was ∼ 1.9 times higher than in the duos. By linkage group, the average difference in LR was 5.5 times higher in favor of the trios; while for the combined LR, the value in trios was 1.036 times higher than in the duos, all of this consistent with the loss of informativeness due to the lack of one of the parents in the duo-type cases.
The typical PI was estimated for each X-STR, an a priori parameter to evaluate the informativeness of autosomal STRs.14 According to the typical PI, DXS8378 (5.3) and HPRTB (4.1) would be the most informative loci to evaluate kinship, as well as the linkage group GL4. This contrasts with the average LR obtained in duos and trios, where DXS7132 and DXS10101 stood out, as well as GL2 and GL3, respectively. In addition, the combined typical PI (a priori) was much lower than the combined average LR (a posteriori) in Mexican families (3.98E+04 < 6.1E+11) (Table 1). Because the inheritance of X-STRs from a couple to daughters conforms to the Mendelian inheritance of autosomal STRs, the typical PI would be expected to be consistent with the informativeness of X-STRs in standard paternity cases. However, the differences observed allow us to conclude that the typical PI underestimates the actual informativeness offered by the X-STRs of the Argus X-12 kit to confirm positive kinship relationships. The high informativeness of X-STRs in paternity cases justifies their inclusion as auxiliary markers in forensic genetics laboratories.
Complex kinship cases resolved with X-STRTo generate an overview of what the estimated LRs in paternity represent, the LR of 4 cases of complex kinship between women resolved with the 12 X-STRs of the Argus X-12 kit is reported (Fig. 1). Recently, 7 cases of identification in the Colombian population were reported where the Mexican database was used to estimate the exact LR.15 However, in most of these, a male or several individuals are involved, that is, they are not analogous to those described here, so only the case of paternal grandmother-granddaughter is discussed as a reference (LR = 1.25E+07). The highest LR was estimated in the case of paternal aunt-niece and half-siblings via the paternal line, both in the presence of their mothers, followed by paternal grandmother-granddaughter, while maternal half-siblings generated the lowest LR. In our experience as a private paternity laboratory, the cases that generated the extreme LRs are those that occur less frequently among those analyzed. On the one hand, the case of the paternal aunt-niece and the paternal half-sibling relationship, both in the presence of the mothers (highest LRs: 2.18E+8 and 6.0E+7, respectively), and on the other hand, the half-sibling relationship via the maternal route (lowest LR: 111.5). For this last case, STRs must be added to generate a conclusive LR.11 One of the main reasons why these cases are not so frequent in the field of paternity is the presence of the mothers, who are not always willing to participate. The case of the paternal aunt-niece relationship is notable because they shared the complete X-STR profile (4/4 haplotypes), which is not the most common between 2 siblings (the paternal aunt and the alleged father who does not participate), according to the binomial probability (p = .0625), but which in this case allowed the aunt-niece relationship to be consistently confirmed (W = 99.9999995%).
Complex kinship cases analyzed with the Argus X-12 kit and the exact LR value estimated using the Mexican population database.9
The paternal grandmother-granddaughter case generated a conclusive LR, but much lower than that described in Colombia (W = 99.994% vs 99.99999%).9 This difference is explained -in part- by the use of a Mexican database with profiles from Colombia, and by the high range of these estimates, as seen in our results for paternity with 12 X-STRs (Table 1).
The LR of the half-siblingship via the paternal line -although discrete- was very close to being conclusive (W = 99.98%). In our experience, both the paternal grandmother-granddaughter and the half-siblingship via the paternal line are the 2 most frequent cases where X-STRs are crucial to resolve them satisfactorily. Regarding the standard paternity values, the average combined LR of the trios (8.0E+14) was -in general- higher than that estimated in the complex cases analyzed (Fig. 1). Also, although the average combined LR of the duos (6.1E+11) was also greater than the LR of the complex cases, the wide range of LR values is striking, since it includes inconclusive values (LR range = 130–5.89E+13) (Table 1).
Mutation rateA low mutation rate of genetic markers used in paternity is essential to avoid false exclusions or low kinship indices or LR.3,8 Until a few years ago, studies to establish the mutation rate of X-STRs were scarce and derived from isolated or individual efforts.4,7 However, this panorama changed with the collaborative study of GHEP-ISFG that brought together 1612 trios (father, daughter and mother), where in addition to adding data available in the literature to achieve a substantial improvement in estimates of mutation rates in X-STRs.8 It is interesting that the possibility of mutation rates varying between populations has been reported poblaciones,1,7 which justifies research on specific mutation rates of various populations, especially those whose biogeographic component has been little studied.
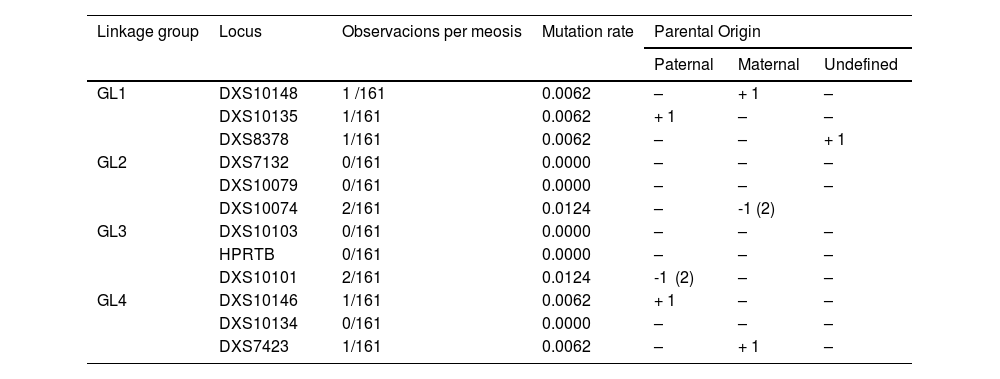
In the 74 Mexican families included in this study, 9 mutations were observed in 161 meiosis (Table 2) where the genotypes and alleles involved can be consulted in Supplementary Material S3. The number of paternal and maternal mutations was the same (n = 4), apart from one mutation whose parental origin could not be defined.9 Although the same proportion of parental origin of mutations is possible (1:1), a higher frequency of paternal mutations has been described, which would make it more likely that the undefined mutation is of paternal origin.8,16 In addition, a similar ratio of gain:loss was observed (5:4), which is in agreement with previous observations.8 The X-STR loci that showed the highest mutation rate were DXS10074 and DXS10101 (μ = 0.0124), followed by DXS10148, DXS10135, DXS8378, DXS10146, and DXS7423 (μ = 0.0062) (Table 2). By linkage group, GL1 showed the highest mutation rate (μ = 0.0186), while the rest had the same mutation rate (μ = 0.0124). It is worth mentioning that the mother-daughter exclusion observed in DXS7423, which involves a 14 → 15 step (Supplementary Material S2), could also be explained by a null allele. The recommended method to confirm the presence of the null allele in DXS7423 would be to re-amplify the sample with another alternate X-STR system,11 assuming that the primers of the alternate kit will align in a different position to achieve amplification of the null allele. Unfortunately, in Mexico, we do not have another commercial kit for X-STR that facilitates this task. However, the expected frequency in the Mexican population of the 14/14 genotype involved in this case is relatively high (p = .0864),17 which makes it plausible that this finding can be explained by mutation. Finally, it is necessary to emphasize that the X-STR mutation rates estimated in this work are not the main contribution of the same, as it was not designed to generate reliable estimates. However, in the literature, we find several studies on mutation rates in which findings - such as those reported here - are integrated into meta-analyses to offer conclusive information.7,8
Mutation rate by X-STR and linkage group (GL) of the Investigator Argus X-12 system estimated from eight mutations found in 74 Mexican families, including the father, mother and daughters.
| Linkage group | Locus | Observacions per meosis | Mutation rate | Parental Origin | ||
|---|---|---|---|---|---|---|
| Paternal | Maternal | Undefined | ||||
| GL1 | DXS10148 | 1 /161 | 0.0062 | – | + 1 | – |
| DXS10135 | 1/161 | 0.0062 | + 1 | – | – | |
| DXS8378 | 1/161 | 0.0062 | – | – | + 1 | |
| GL2 | DXS7132 | 0/161 | 0.0000 | – | – | – |
| DXS10079 | 0/161 | 0.0000 | – | – | – | |
| DXS10074 | 2/161 | 0.0124 | – | -1 (2) | ||
| GL3 | DXS10103 | 0/161 | 0.0000 | – | – | – |
| HPRTB | 0/161 | 0.0000 | – | – | – | |
| DXS10101 | 2/161 | 0.0124 | -1 (2) | – | – | |
| GL4 | DXS10146 | 1/161 | 0.0062 | + 1 | – | – |
| DXS10134 | 0/161 | 0.0000 | – | – | – | |
| DXS7423 | 1/161 | 0.0062 | – | + 1 | – | |
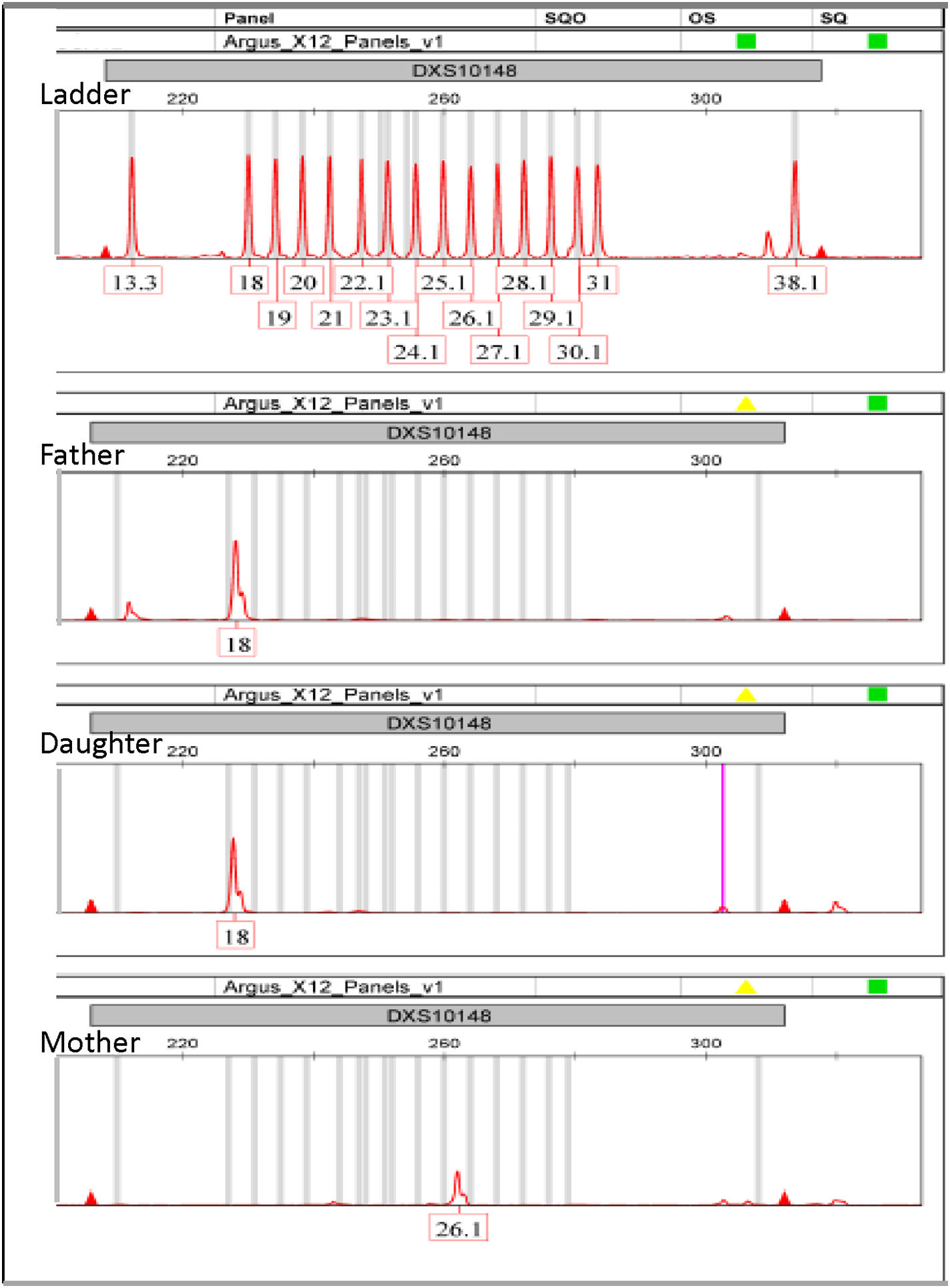
Samples from the families analyzed were amplified with the Argus X-12 QS system because, according to the commercial company, this HID kit includes additional primers that prevent the high-frequency appearance of null alleles in DXS10101, DXS10146 and DXS10148, observed in African populations.18 In one of the families, an isolated exclusion between mother and daughter was observed for DXS10148, where the mother appears to be a 26.1/26.1 homozygote while the father and daughter share the 18 alleles (Fig. 2). This could be explained either by a maternal 26.1 → 18 mutation or by maternal inheritance of a null allele, also known as a silent allele.
Regarding the mutation hypothesis, according to the single-step mutation model, known as SSM, an 8-step maternal mutation (26.1 → 18) would be very unlikely, since it has been reported that 90–98 % of the time mutations involve only one repeat (+1 or −1) and most are of paternal origin.19,20 In particular, the most extensive study on X-STR mutations with the Argus X-12 kit describes single-step mutations in 96.1 % (174/181), and 2-step mutations in 2.7 %, without having found mutations with more than 2 steps where -in addition- paternal mutations are 5.2 times more frequent than maternal ones.8 For this case, one of the most widely used ways of estimating mutations with more than 2 steps is that of Brenner,21 who indicates that the probability of mutation is approximately 10 % of each step, taking into account that approximately half are gains and the other half are losses. The formula for the probability of mutation in this case, considering that the mother would be “homozygous” and could only have inherited one allele, is the following: μ (1/2)(1/10)s−1; where s is the number of steps. Applying the estimated maternal mutation rate for DXS10148 (1.83 E−03)8 and that the change 26.1 → 18 would imply 8 steps, the probability of this mutation would be 9.15E−05. As an additional fact, the expected low frequency of homozygous 26.1 in the Mexican population (0.014)),17 reduces the possibility of the hypothesis of mutation in a homozygous 26.1 woman.
Furthermore, the presence of a null allele generally implies a mutation at the site where the primer binds, so it is recommended to use an additional pair of primers, usually applying a different STR kit to reveal that it is a null allele.19 The frequency of these alleles depends on the STR kit used, the genetic marker and the population involved, so commercial companies have made efforts to modify their kits by applying additional primers or modifying them completely so that they hybridize at another site to thus decrease the frequency of null alleles.18,22 Unfortunately, in our laboratory –and in Mexico– we do not have another commercial kit for X-STR that allows us to confirm this finding. In short, for this case, the low probability of the mutation (9.15E-05), in a rare genotype in this population (0.014), where mutations are usually paternal and not maternal (5:1), practically allows us to rule out the possibility of mutation, while the hypothesis of a null allele is the most plausible.
ConclusionsIn short, from Mexican families with daughters and proven paternity, different a posteriori parameters were estimated that describe the informativeness of the Investigator Argus X-12 kit to establish standard paternity relationships. Some complex cases of complex kinship resolved with X-STR are described -as a reference. From the findings, the mutation rate was estimated for 7 X-STRs and a probable null allele in DXS10148.
Ethical responsibilitiesThe participants signed an informed consent letter approved according to the Declaration of Helsinki. The project was approved by the Research Ethics Committee of the Institute of Molecular Genetics Research of the CU Ciénega, University of Guadalajara. The anonymity of the volunteers included in this study was preserved at all times.
FundingCONACyT-Mexico for funding N°286623 to H. R-V and scholarships to E. R-P, and postdoctoral fellowship to M.E. G-A (N°664279). QIAGEN-Mexico has partially supported this project.
AuthorshipEduardo Rojas-Prado, Irán Cortés-Trujillo and Mayra E. García-Aceves equally contributed to this study.
Our sincere thanks to the families who voluntarily supported this project with their samples.
Please cite this article as: Rojas-Prado E, Cortés-Trujillo I, García-Aceves ME, Chávez-Arreguín M, González-Becerra K, Favela Mendoza AF, Rangel-Villalobos H. A posteriori informativity in standard paternity tests with the Investigator Argus X-12 system and mutation findings. Revista Española de Medicina Legal. 2025. https://doi.org/10.1016/j.remle.2025.100426.