Molecular markers in leukaemia are essential to diagnose, establish prognosis factors and determine the correct treatment of patients; therefore, it is imperative to include molecular biology studies, so that, combined with cytomorphology and immunophenotyping studies, they constitute the differential diagnosis of these neoplasias. It is extremely important to implement a panel of molecular markers that allows us to detect oncogenes derived from chromosomal translocations, genes derived from epigenetic alterations and drug-resistant genes.
A panel of molecular markers that included 11 genes derived from chromosomal translocations BCR-ABL major and minor breakpoints, E2A-PBX1, MLL-AF4, TEL-AML1, PML-RARα, AML1-ETO was standardised; cancer testis antigens (CTA) derived from NY-ESO1 and MAGE-A3 epigenetic alterations and multi-drug-resistant genes ABCB1 and ABCG2. 30 patients diagnosed with leukaemia from Mexico's General Hospital (Hospital General de Mexico) were included. They suffered from acute lymphoblastic leukaemia (ALL) and acute myeloblastic leukaemia (AML); bone marrow mononuclear cells were used, from which RNA was extracted for the synthesis of cDNA and RT-PCR for each of the markers. In acute lymphoblastic leukaemia (ALL), BCR-ABL biomarkers expressed under 30% (3/10), E2A-PBX1 10% (1/10), ABC-B1 80% (8/10), and ABC-G2 60% (6/10). Patients with acute myeloblastic leukaemia (AML) expressed 30% PML-RARα (3/10), 40% ABC-B1 (4/10), and 10% ABC-G2 (1/10). Lastly, in patients with chronic myeloid leukaemia (CML), BCR-ABL was over 100% (10/10), ABC-B1 20% (2/10), and ABC-G2 50% (5/10). The presence of transcripts from chimeric genes minor BCR-ABL and E2A-PBX1 in ALL; PML-RARα in AML; and major BCR-ABL in CML, confirms the importance that the panel of molecular markers has in strengthening the diagnosis and prognosis of these conditions.
Los marcadores moleculares en leucemias son fundamentales para el diagnóstico, el establecimiento de factores pronósticos y la determinación del tratamiento adecuado para los pacientes; por tanto es obligado incluir los estudios de biología molecular, para que aunado a los de citomorfología e Inmunofenotipo, conformen el diagnóstico diferencial de estas neoplasias. Es de gran importancia implementar un panel de marcadores moleculares que permita detectar oncogenes derivados de translocaciones cromosómicas, genes derivados de cambios epigenéticos y de resistencia genes a drogas.
Se estandarizó un panel de marcadores moleculares que incluyeron 11 genes derivados de translocaciones cromosómicas BCR-ABL rompimiento mayor y menor, E2A-PBX1, MLL-AF4, TEL-AML1, PML-RARα, AML1-ETO; los antígenos testiculares de cáncer (ATC) derivados de cambios epigenéticos NY-ESO1 y MAGE-A3 y genes de resistencia multidrogas ABCB1 y ABCG2. Se incluyeron 30 pacientes con diagnóstico de leucemia del Hospital General de México, de los tipos, leucemia aguda linfoblástica (LAL) y mieloblástica (LAM) así como leucemia mieloide crónica (LMC); se utilizaron células mononucleares de médula ósea, a las cuales se les extrajo RNA para la posterior síntesis de cDNA y RT-PCR para cada uno de los marcadores. En leucemia linfoblástica aguda (LAL) se expresaron los biomarcadores BCR-ABL menor 30% (3/10), E2A-PBX1 10% (1/10), ABC-B1 80% (8/10), y ABC-G2 60% (6/10). Los pacientes con leucemia mieloblástica aguda (LAM) expresaron PML-RARα 30% (3/10), ABC-B1 40% (4/10), y ABC-G2 10% (1/10). Finalmente, en pacientes con leucemia mieloide crónica (LMC) se encontró BCR-ABL mayor 100% (10/10), ABC-B1 20% (2/10), y ABC-G2 50% (5/10). La presencia de los transcritos de los genes quiméricos BCR-ABL menor y E2A-PBX1 en LAL; PML-RARα en LAM; y BCR-ABL mayor en LMC, confirma la importancia del panel de marcadores moleculares en el fortalecimiento del diagnóstico y pronóstico para dichos padecimientos.
Over the last few years, there has been major progress in the understanding of the molecular mechanism associated with normal haematopoiesis and with the development of haematological neoplasias.1,2 The molecular alterations in genes that control cell differentiation programs, such as proto-oncogenes and tumour suppressor genes, derive in the loss of homeostasis regulation in haematopoietic tissue and promote the development of leukaemia.3 There are currently over 50 different chromosomal alterations associated with leukaemia.4–6 The most common damage mechanisms are the balanced chromosomal translocations. A number of times, there are genes involved in chromosomal translocations in which the products are transcription factors that control differentiation mechanisms in haematopoietic tissue precursor cells.5–7 These transcription factors are responsible for the malignant transformation, and they are essential elements in the design of alternative treatment plans.8,9 Other molecular alterations that take place in the development of leukaemia are those called point mutations, which damage the mechanisms that regulate cell proliferation, apoptosis and differentiation in haematopoietic precursors.10,11 Lastly, there are additional alterations that do not affect the information held in the DNA nucleotide sequence, which are associated with epigenetic events; that is to say, they lead to DNA hypermethylation or to abnormal histone acetylation, affecting the transcriptional availability of proto-oncogenes and suppressor genes.12,13 Mexico does not have a tumour marker panel that could be used to perform the diagnosis, prognosis and follow-up for leukaemia, both acute and chronic, by means of molecular methods such as the RT-PCR. Our laboratory analysed the expression of 11 tumour markers in mononuclear cells from 30 patients with leukaemia. The genes under analysis are involved in the proliferation, differentiation, epigenetic events and resistance mechanisms to drugs. Among the genes derived from translocations are: BCR-ABL major and minor breakpoint, t(9;22), E2A-PBX1, t(1;19), TEL-AML1 t(12;21), AF4-MLL t(4;11), AML1-ETO, t(8;21),14–16 alterations that take place due to changes in the methylation pattern, such as the testicular cancer genes that are only present in tumour cells and absent in normal tissue; MAGE-A3, NY-ESO, MAGE-4,17,18 and genes that express as resistance mechanisms to multiple drugs, such as ABC-B1 (MDR-1) and ABC-G2 (BCR-P) genes.19,20
The results showed expression of the tumour markers in leukaemia and their potential use for the diagnosis and prognosis of these oncohaematologic diseases.
MethodologyAn experimental, prospective study was carried out between February and December, 2014, at the Molecular and Cell Biology Department of the Haematology Service. The study was authorised by the Institution's Research and Ethics Committees. 30 patients of both genders with ALL, AML and de novo CML were included in the study. Bone marrow aspiration was performed after informed consent was signed.
Mononuclear cell extractionMononuclear cells were extracted using Lymphoprep (NycomedPharma AS, Oslo, Norway) according to the supplier's instructions and stored at −80°C until they were used.
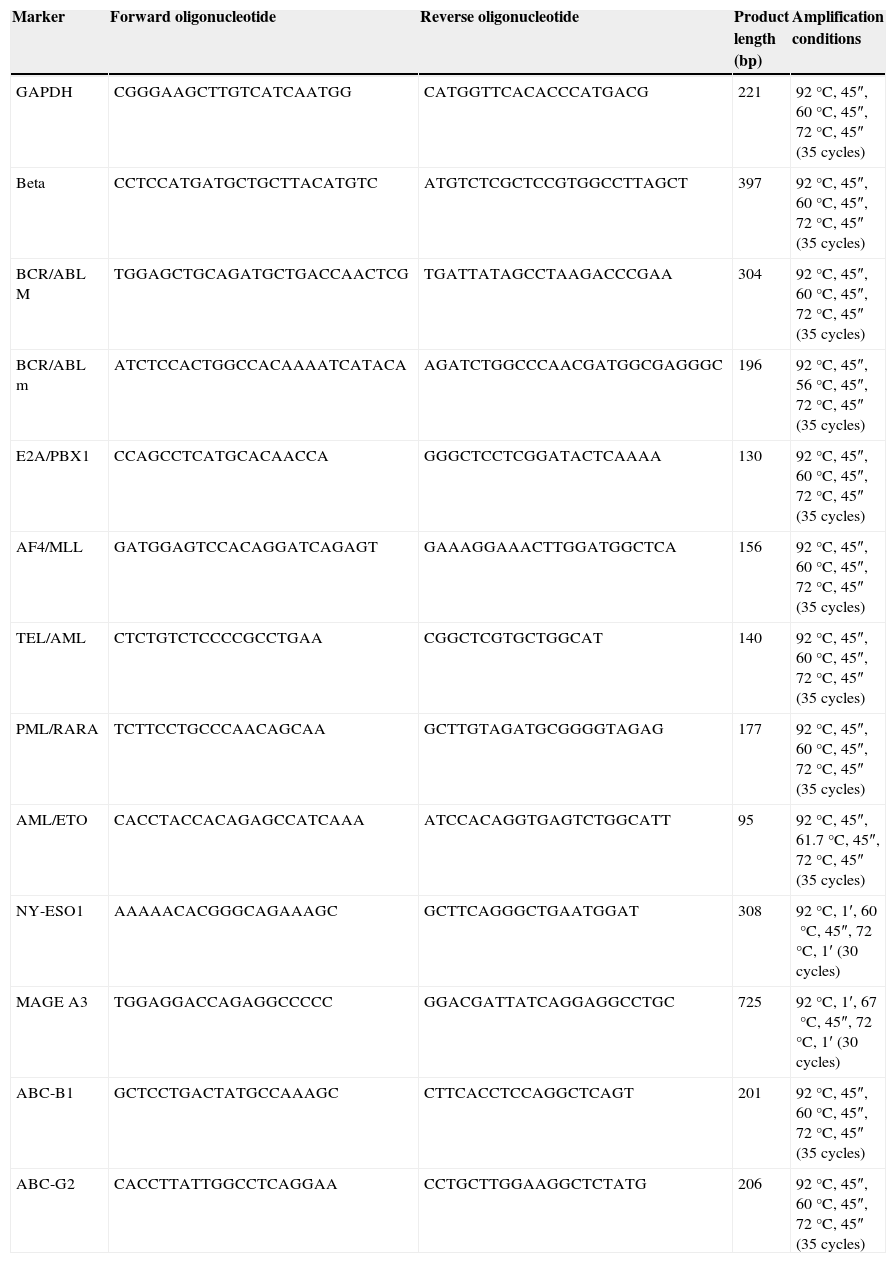
RT-PCRTotal cell RNA extraction was performed through Trizol reactive (Life Technologies, UK); 5μg of RNA was used to synthesise cDNA through MMLV (Life Technologies, Paisley, UK). CRP amplification was performed using specific primers (Table 1).
Sequence of the primers used and their thermal profiles.
| Marker | Forward oligonucleotide | Reverse oligonucleotide | Product length (bp) | Amplification conditions |
|---|---|---|---|---|
| GAPDH | CGGGAAGCTTGTCATCAATGG | CATGGTTCACACCCATGACG | 221 | 92°C, 45″, 60°C, 45″, 72°C, 45″ (35 cycles) |
| Beta | CCTCCATGATGCTGCTTACATGTC | ATGTCTCGCTCCGTGGCCTTAGCT | 397 | 92°C, 45″, 60°C, 45″, 72°C, 45″ (35 cycles) |
| BCR/ABL M | TGGAGCTGCAGATGCTGACCAACTCG | TGATTATAGCCTAAGACCCGAA | 304 | 92°C, 45″, 60°C, 45″, 72°C, 45″ (35 cycles) |
| BCR/ABL m | ATCTCCACTGGCCACAAAATCATACA | AGATCTGGCCCAACGATGGCGAGGGC | 196 | 92°C, 45″, 56°C, 45″, 72°C, 45″ (35 cycles) |
| E2A/PBX1 | CCAGCCTCATGCACAACCA | GGGCTCCTCGGATACTCAAAA | 130 | 92°C, 45″, 60°C, 45″, 72°C, 45″ (35 cycles) |
| AF4/MLL | GATGGAGTCCACAGGATCAGAGT | GAAAGGAAACTTGGATGGCTCA | 156 | 92°C, 45″, 60°C, 45″, 72°C, 45″ (35 cycles) |
| TEL/AML | CTCTGTCTCCCCGCCTGAA | CGGCTCGTGCTGGCAT | 140 | 92°C, 45″, 60°C, 45″, 72°C, 45″ (35 cycles) |
| PML/RARA | TCTTCCTGCCCAACAGCAA | GCTTGTAGATGCGGGGTAGAG | 177 | 92°C, 45″, 60°C, 45″, 72°C, 45″ (35 cycles) |
| AML/ETO | CACCTACCACAGAGCCATCAAA | ATCCACAGGTGAGTCTGGCATT | 95 | 92°C, 45″, 61.7°C, 45″, 72°C, 45″ (35 cycles) |
| NY-ESO1 | AAAAACACGGGCAGAAAGC | GCTTCAGGGCTGAATGGAT | 308 | 92°C, 1′, 60°C, 45″, 72°C, 1′ (30 cycles) |
| MAGE A3 | TGGAGGACCAGAGGCCCCC | GGACGATTATCAGGAGGCCTGC | 725 | 92°C, 1′, 67°C, 45″, 72°C, 1′ (30 cycles) |
| ABC-B1 | GCTCCTGACTATGCCAAAGC | CTTCACCTCCAGGCTCAGT | 201 | 92°C, 45″, 60°C, 45″, 72°C, 45″ (35 cycles) |
| ABC-G2 | CACCTTATTGGCCTCAGGAA | CCTGCTTGGAAGGCTCTATG | 206 | 92°C, 45″, 60°C, 45″, 72°C, 45″ (35 cycles) |
Each cDNA was tested by CRP using specific primers for two genes of constitutive expression β2 microglobulin and GADPH. CRP reaction was carried out according to the alignment temperature previously shown in Table 1. CRP products were visualised in 2% agarose gel dyed with Midori Green Advanced (Nippon Genetics, Germany).
The amplified segments were sequenced using the ABI Prism Dye Terminator Cycle Sequencing kit. The alignment was performed using the Gene-Bank BLASTN program, generating a 99% similarity with all the biomarkers used.
ResultsCharacteristics of the patientsIn total, 30 patients were under study; 10 of each type (ALL, AML, CML), the median age for each group was of 32 for ALL and 34 for AML and CML. Ages ranged between 20–55 years, 21–68 years and 18–50 years, respectively.
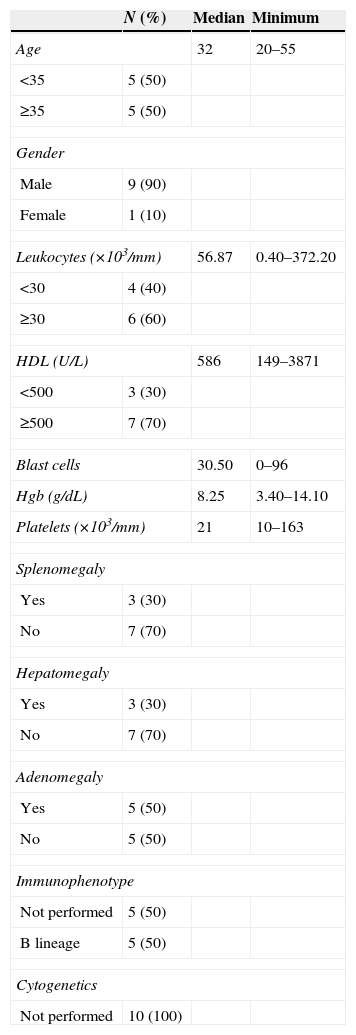
Regarding ALL, the morphologic classification corresponded to type L2, 70% had a B-cell immunophenotype. Only three patients had hepatomegaly and one of them had high levels of HDL, hyperleukocytosis, low levels of platelets (Table 2).
Characteristics of the patients with ALL.
| N (%) | Median | Minimum | |
|---|---|---|---|
| Age | 32 | 20–55 | |
| <35 | 5 (50) | ||
| ≥35 | 5 (50) | ||
| Gender | |||
| Male | 9 (90) | ||
| Female | 1 (10) | ||
| Leukocytes (×103/mm) | 56.87 | 0.40–372.20 | |
| <30 | 4 (40) | ||
| ≥30 | 6 (60) | ||
| HDL (U/L) | 586 | 149–3871 | |
| <500 | 3 (30) | ||
| ≥500 | 7 (70) | ||
| Blast cells | 30.50 | 0–96 | |
| Hgb (g/dL) | 8.25 | 3.40–14.10 | |
| Platelets (×103/mm) | 21 | 10–163 | |
| Splenomegaly | |||
| Yes | 3 (30) | ||
| No | 7 (70) | ||
| Hepatomegaly | |||
| Yes | 3 (30) | ||
| No | 7 (70) | ||
| Adenomegaly | |||
| Yes | 5 (50) | ||
| No | 5 (50) | ||
| Immunophenotype | |||
| Not performed | 5 (50) | ||
| B lineage | 5 (50) | ||
| Cytogenetics | |||
| Not performed | 10 (100) | ||
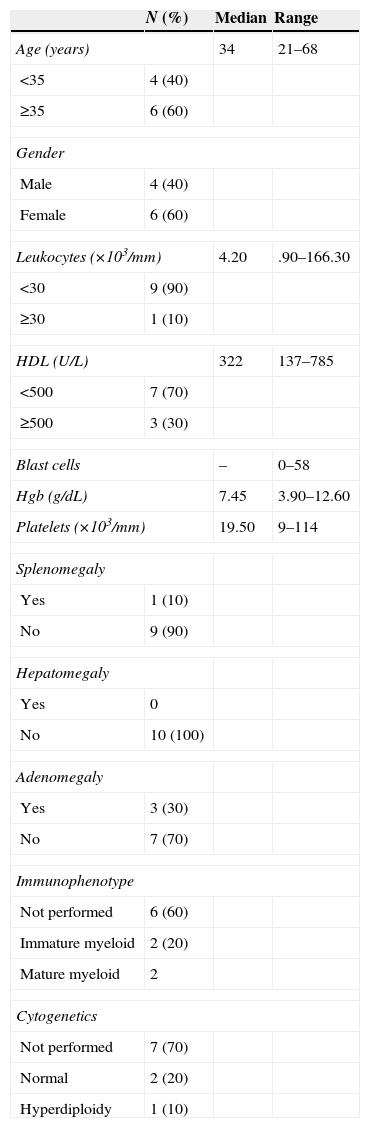
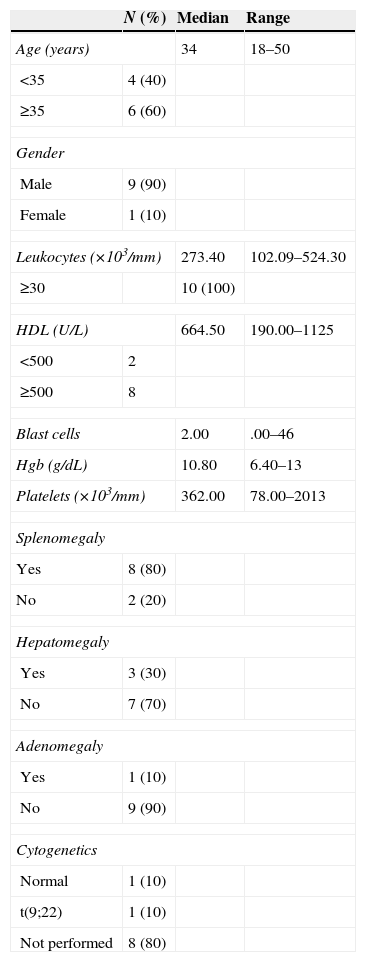
In the group of patients with AML; five patients were AML-M3 (acute promyelocytic leukaemia), four were AML-M4 (acute myelomonocytic leukaemia) and one was AML-M2 (acute myeloid leukaemia with maturation). 60% of the patients were over the age of 60 and only one reported having hyperleukocytosis. One patient had splenomegaly and three patients had adenomegaly. As for the cytogenetic alterations, only one patient had hyperdiploidy (Table 3). Lastly, out of the patients with CML, 60% was above the age of 35, 100% had hyperleukocytosis, and eight patients had high levels of HDL. 80% had splenomegaly, 10% had hepatomegaly and only 10% had adenomegaly. The cytogenetic alterations reported had t(9;22) only among 10% of the population and the study was not performed in 90% of the population (Table 4).
Characteristics of the patients with AML.
| N (%) | Median | Range | |
|---|---|---|---|
| Age (years) | 34 | 21–68 | |
| <35 | 4 (40) | ||
| ≥35 | 6 (60) | ||
| Gender | |||
| Male | 4 (40) | ||
| Female | 6 (60) | ||
| Leukocytes (×103/mm) | 4.20 | .90–166.30 | |
| <30 | 9 (90) | ||
| ≥30 | 1 (10) | ||
| HDL (U/L) | 322 | 137–785 | |
| <500 | 7 (70) | ||
| ≥500 | 3 (30) | ||
| Blast cells | – | 0–58 | |
| Hgb (g/dL) | 7.45 | 3.90–12.60 | |
| Platelets (×103/mm) | 19.50 | 9–114 | |
| Splenomegaly | |||
| Yes | 1 (10) | ||
| No | 9 (90) | ||
| Hepatomegaly | |||
| Yes | 0 | ||
| No | 10 (100) | ||
| Adenomegaly | |||
| Yes | 3 (30) | ||
| No | 7 (70) | ||
| Immunophenotype | |||
| Not performed | 6 (60) | ||
| Immature myeloid | 2 (20) | ||
| Mature myeloid | 2 | ||
| Cytogenetics | |||
| Not performed | 7 (70) | ||
| Normal | 2 (20) | ||
| Hyperdiploidy | 1 (10) | ||
Characteristics of the patients with CML.
| N (%) | Median | Range | |
|---|---|---|---|
| Age (years) | 34 | 18–50 | |
| <35 | 4 (40) | ||
| ≥35 | 6 (60) | ||
| Gender | |||
| Male | 9 (90) | ||
| Female | 1 (10) | ||
| Leukocytes (×103/mm) | 273.40 | 102.09–524.30 | |
| ≥30 | 10 (100) | ||
| HDL (U/L) | 664.50 | 190.00–1125 | |
| <500 | 2 | ||
| ≥500 | 8 | ||
| Blast cells | 2.00 | .00–46 | |
| Hgb (g/dL) | 10.80 | 6.40–13 | |
| Platelets (×103/mm) | 362.00 | 78.00–2013 | |
| Splenomegaly | |||
| Yes | 8 (80) | ||
| No | 2 (20) | ||
| Hepatomegaly | |||
| Yes | 3 (30) | ||
| No | 7 (70) | ||
| Adenomegaly | |||
| Yes | 1 (10) | ||
| No | 9 (90) | ||
| Cytogenetics | |||
| Normal | 1 (10) | ||
| t(9;22) | 1 (10) | ||
| Not performed | 8 (80) | ||
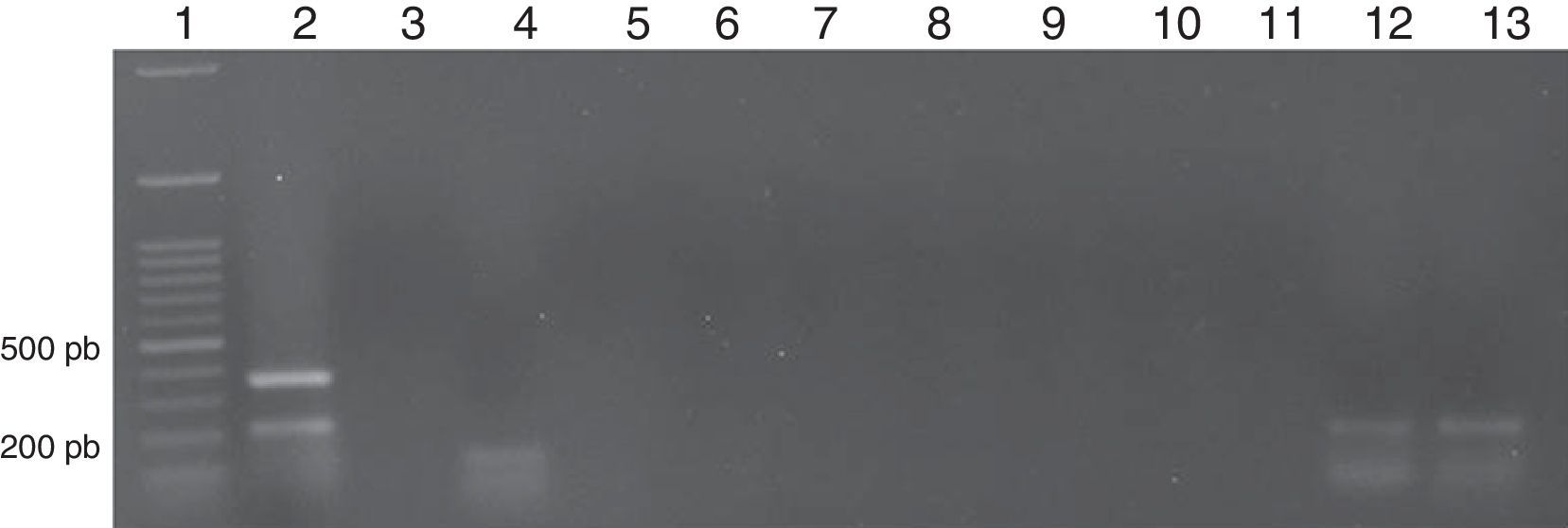
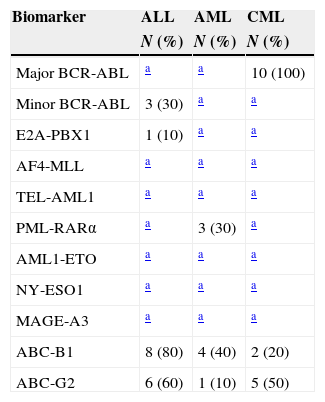
The expression of 11 cancer biomarkers was analysed (BCR-ABL major, minor, ABC-B1, ABC-G2, E2A-PBX1, AF4-MLL, TEL-AML1, PML-RAR alpha, AML1-ETO, NY-ESO, MAGE-A3) via RT-PCR on end-point in patients with ALL, AML and CML. The quality of the RNA was evaluated by means of the amplification of the genes of constitutive expression β2 microglobulin that amplifies an expected fragment of 397bp and GADPH by amplifying a 221bp fragment. The markers expressed in ALL were: BCR-ABL minor breakpoint with an amplification product of 196bp, detected in 30% (3/10), ABC-B1 by amplifying a 201bp fragment in 80% (8/10) and ABC-G2 with a 206bp fragment in 60% (6/10). In addition, the expression of oncogene E2A-PBX1 with a product of 130bp was detected in 10% (1/10) (Fig. 1).
Representative image of a patient with LAL. Lane 1: Molecular weight marker; Lane 2: β-2-microglobulin (394 pb) and GAPDH (221 pb); Lane 3: BCR-ABL M (−); Lane 4: BCR-ABL m (150 pb); Lane 5: E2A-PBX1 (−); Lane 6: TEL-AML1 (−); Lane 7: AF4-MLL1 (−); Lane 8: PML-RARα (−); Lane 9: AML1-ETO (−); Lane10: MAGE-A3 (−); Lane 11: NY-ESO1 (−); Lane 12: ABC-B1 (201 pb); Lane13: ABC-G2 (206 pb).
In the case of patients with AML, the biomarkers identified were: ABC-B1 in 40% (4/10), ABC-G2 in 10% (1/10) and PML-RAR alpha with an amplified product of 177bp in 30% (3/10). Lastly, the expression of biomarkers detected in CML was BCR-ABL major breakpoint with an amplification fragment of 304bp in 100% (10/10), ABC-B1 in 20% (2/10) and ABC-G2 in 50% (5/10) (Table 5).
Frequency of the panel of biomarkers in leukaemia.
In the last 20 years, several molecular methods have been used to detect genetic abnormalities that are important for the diagnosis and prognosis of leukaemias.21,22 Early detection is essential to make decisions about specific treatments, such as with tyrosine kinase inhibitors (Imatinib, Nilotinib or Dasatinib), used in patients with CML and ALL who express BCR-ABL oncogenic protein, as well as the use of trans-retinoic acid used in patients with AML-M3 who express PML-RAR alpha protein. These treatments have shown a survival increment of up to 80% in five years.23,24 We searched for seven tumour markers derived from translocations, BCR-ABL major and minor, E2A-PBX1, AF4-MLL, TEL-AML1, PML-RAR alpha and AML1-ETO. The results showed the expression of BCR-ABL minor breakpoint in 30% of ALL and E2A-PBX1 expression in 10%; these results correspond to what was reported previously.25,26 The detection of BCR-ABL in patients with ALL enables us to select the use of inhibitors that revert the development of the disease.27 Regarding AMLs, the expression of oncogene PML-RAR alpha was detected in 30% of the AML-M3, which coincides with the international series. Its detection is essential to begin treatment with trans-retinoic acid.28 It is important to point out that only one AML-M3 case had the t(15;17) by cytogenetics, from which the PML-RAR alpha transcript is derived. In the case of CMLs, only the BCR-ABL major breakpoint expression was observed in 100%, which coincides with what was reported previously. This marker is fundamental to begin treatment with Imatinib or Nilotinib, and it increases survival in up to 80% in five years.29,30 Our panel confirms the clinical diagnosis was performed in all cases. Cell resistance to chemotherapy drugs can represent the main cause of failure of current treatments.31,32 As for the expression of drug-resistant genes, ALLs express ABC-B1 more frequently, followed by AMLs and CMLs. In the case of ABC-G2, the expression is higher in ALLs, followed by CMLs and AMLs. It has been reported that the percentage of healing cases in ALLs in five years is of 30% and in AML it is between 15 and 25%.33 Olarte Carrillo et al. reported an ABC-B1 frequency of expression of 27.3% in patients with AML. As for ALL, the frequency varies between 22 and 47% by protein analysis.34,35 The expression at the time of diagnosis could be of use during the follow-up treatment and prognosis of the disease. There is controversy over its predictive association. Our research group previously reported an association with treatment failure in 22 patients with AML.36,37 In the case of the expression of genes ABC-G2, only an increment in malignant progenitor cells derived from CML has been reported.38 In this study, an increment in the frequency of CML and ALL was found, so this could be associated with the mechanisms of resistance to inhibitors. In the case of the biomarkers involved in epigenetic changes like MAGE A3 and NY-ESO-1, there was no sign of expression in any of the analysed samples. Previously, in a series of 115 patients, MAGE-A3 expression was found in 41% of AML and 30% of ALL,39 involved in the progression of the disease; in the case of MAGE A3, we have detected its expression in 46% of diffuse large B-cell lymphomas and an involvement with a lower survival rate.17,40
In conclusion, the panel used by means of RT-PCR enables to back routine methods for decision-making in treatments, not only in the beginning but also in the detection of the minimal residual disease, which enables assessment of the tumour load in a precise and specific manner. Having a panel of 11 standardised markers reduces the time it takes to arrive at a diagnosis of oncohaematologic diseases. Knowing the frequency of expression within our population enables us to have timely detection and control, which improves the performance of current available treatments.
Conflict of interestThe authors declare that they have no conflict of interests.
This work was supported by Consejo Nacional de Ciencia y Tecnologia, CONACYT, registered under project number 162269 and 80085, as well as by the Research Department from Mexico's General Hospital under registry numbers DIC/09/04/03/131, DIC/08/204/04/017, DIC/12/204/05/01 and Novartis Oncology: CSTI571AMX10T