Despite the advances in investigative techniques in Forensic Genetics, there are still a large number of unsolved criminal cases. We have recently seen the birth of a new forensic discipline called forensic genealogy or investigative genetic genealogy, which allows to us resolve satisfactorily many of these cold cases combining DNA analysis technology with genealogical search tools.
The potential and risks of this new research strategy are manifest. We must maintain a balance between personal privacy and personal interests on the one hand and on the other, public security, the good of the community and the resolution and clarification of criminal acts, requiring a social, legal and scientific debate that clarifies all these aspects and enables adequate legal regulation of these practices.
A pesar del avance de las técnicas investigativas de Genética Forense, todavía existen gran cantidad de casos criminales no resueltos y en muchos casos inabordables por su antigüedad. Recientemente hemos asistido al nacimiento de una nueva disciplina forense, la genealogía forense o genealogía genética investigativa, que permite que muchos de estos casos puedan ser resueltos satisfactoriamente combinando la tecnología de análisis de ADN y las herramientas de búsquedas genealógicas.
La potencialidad de esta nueva estrategia investigativa es evidente, de la misma manera que lo son sus riesgos. Hemos de mantener un equilibrio entre la privacidad personal y los intereses de las personas afectadas por un lado y la seguridad pública, el bien de la comunidad y la resolución y esclarecimiento de hechos delictivos por el otro, siendo necesario un debate social, legal y científico que clarifique todos estos aspectos y posibilite una adecuada regulación legal de estas prácticas.
The use of DNA analysis to solve crimes, polymerase chain reaction (PCR), the use of automatic sequencers and the comparison of genetic profiles in international databases have all revolutionised criminal investigation, making it possible to solve thousands of cases. Progress has been made recently in improving DNA extraction, while analysis has been automated, single nucleotide polymorphisms (SNPs) are now used to analyse ancestry-informative and phenotype markers, massive parallel sequencing (MPS) is now used, and software has been developed that makes it possible to interpret complex DNA samples from several contributors.1–4 It used to be hard to imagine that a new revolution could swiftly occur that would enable us to resolve questions that used to be unanswerable, even using the analytical methodology and techniques described above. Nevertheless, all of this changed on 24 April 2018 when the Police arrested Joseph James DeAngelo (known as the “Golden State killer”) in the United States. This retired policeman was guilty of 13 murders, more than 50 rapes and more than 100 robberies. He was caught thanks to the use of a new investigative tool: genealogical genetic identification, or forensic genealogy.5
Before commencing this review, we have to differentiate between DNA databases created for the purpose of criminal investigation and those which have genealogical purposes: the former are governed by the law and contain genetic profiles obtained within the context of criminal investigations, the identification of corpses or people who have disappeared; the latter type lack any legal regulation and contain voluntarily supplied genetic profiles with the aim of seeking potential family members, to complete family trees or to discover ancestors. Supplementary file 1 of Appendix B contains a detailed differentiation between both types of database.
MethodologyForensic genealogy may be defined as the practice of finding family members of an individual suspected of committing a crime, by checking crime scene DNA against family tree databases. It therefore uses genealogical methods in cases with legal implications.
The method involves extracting DNA from a crime scene sample and creating a file with hundreds of thousands (from 600,000 to 900,000) SNPs (the nomenclature and position of these genetic markers are shown in https://my.pgp-hms.org). This file is entered into genealogical databases (fundamentally GEDMatch and FamilyTreeDNA) which hold more than one million DNA files uploaded voluntarily by amateur genealogists who are trying to find a biological parent or relative lost a long time beforehand. The GEDMatch and FamilyTreeDNA algorithms search the database for shared segments of DNA (genetic ligament) and add them. The more the DNA that is shared, the closer the relationship.
By using public registries such as the census, birth and death certificates, newspaper cuttings and social networks, etc., genealogists create family networks that may include thousands of people. They then reduce the list to a smaller group of suspects which they pass to the investigators. These lists of relations generated by shared DNA are treated as clues which the police investigate in greater depth using conventional detective work. A database search generates one or more family trees to create a group of possible candidates. Traditional investigation methods then reduce this group of candidates using data such as location, sex, age and crime scene access, etc. As the relations will often be located on both sides of the family tree of the owner of the crime scene profile, the point where the family trees converge is a critical finding. This indicates that the DNA from both the maternal and paternal sides of the family is represented by a single person or in a single family, allowing specific candidates to be identified for sampling. Reference samples can then be obtained from these candidates for direct comparison with the crime scene profile, for direct inclusion or exclusion.6
There is a clear difference between familial searching in criminal databases7 and forensic genealogy. This difference arises on the one hand in terms of the number of genetic markers (20–30 STRs [short tandem repeats] vs more than 600,000 SNPs), while on the other hand, different databases are used (CODIS vs. GEDmatch, for example). Finally, the degree of relationship which corresponds to success also differs, with parents, children and brothers on the one hand and third cousins on the other, for example.
UsesThe main uses of this new investigative tool include:
- •
The identification of criminals using samples of their DNA left at the crime scene.
- •
The identification of cadaveric remains.
- •
The exoneration of wrongly charged innocent people.
- •
Family reunifications in wartime, etc.
- •
The discovery of unknown family members.
- •
The discovery of genetic relationships between parents.
- •
The discovery of the bio-geographical origin of family members and ancestors.
The human genome has two DNA chains, each one of which contains approximately 3,272 million nucleotides.8 Degree of genetic proximity is measured in centiMorgan (cM) units (1cM is equivalent to 1% probability that a marker in a chromosome will be separated from a second marker in the same chromosome due to the intercrossing phenomenon in a single generation); 1cM is approximately equivalent to one million DNA nucleotides in the human genome.9 We can therefore expect that two univiteline twins who share a complete genome will have about 6,500cM concordance respecting each one of their progenitors or a child that shares 50% of the genome (IBD, identical by descent) will have a concordance of approximately 3,250cM. Obviously the farther the degree of relationship, the lower the amount of shared DNA, so that for example, second cousins who share about 3.12% of the genome will have about 212cM in common.
The degree of DNA shared by two individuals may be measured using three different approaches:10
- •
A probabilistic approach: the probability of finding the same DNA fragments in two individuals based on a specific hypothesis of their degree of relationship. The closer their relationship, the higher the probability.11 We have to consider a certain degree of increased shared DNA as the result of imbalanced binding (the difference between the observed frequency of a combination of alleles and the expected frequency of the same if the association between them were independent and random) so that we are able to use corrector mechanisms to set a threshold of 0.15cM for distances between two markers and exclude markers that are not very polymorphic.10
- •
The KING approach (relationship-based inference for whole genome association studies).12 This counts the number of shared and identical-by-state (IBS) alleles between two individuals in a large number of genetic markers to thereby measure degree of relationship.
- •
The segment approach: this measures the amount (in cM) of the segment of each chromosome that two individuals share.13,14 The length in cM of the shared segment will give us an idea of their degree of relationship. As DNA is transmitted randomly down through generations, the probability of distant family members sharing the same IBD segments will be inversely proportional to the number of generations separating the said individuals.
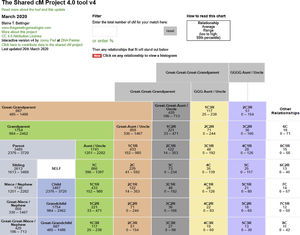
The latter (segment) approach is used by genealogical databases. Fig. 1 shows the shared values in cM between distant family relations.15
DNA segments shared between family members. (Relationships have been eliminated for only one parent or half-sibling, etc.- to improve visualisation). Data obtained from analysis of more than 60,000 known relationships. It is possible to enter a specific percentage value of shared segment between two individuals or a specific percentage of kinship between two individuals, to obtain a probabilistic estimation of their degree of relationship (pedigrees are assumed not to contain any incestuous relationships). For each kinship relationship it is possible to display a histogram showing the distribution of the number of segments shared in the relationship in question.
(https://dnapainter.com/tools/sharedcmv4) (Accessed 13 May 2020).
Companies do not all analyse the same number of SNPs, and sometimes a sample containing little or degraded DNA will not have maximum efficacy. Sometimes it is only possible to have 20% of markers in common to establish coincidences. A method known as genetic imputation is used to resolve this problem.16 This method consists of inferring DNA results which are common in different populations but which have not been tested directly in the user’s DNA. In other words, the absent DNA is predicted (imputed) based on the DNA in adjacent locations, as normally both are found in association.
Imputation is based on two premises:
- •
DNA locations are generally inherited together in groups, in a process known as binding imbalance.
- •
Individuals in common populations share a significant amount of the same DNA.
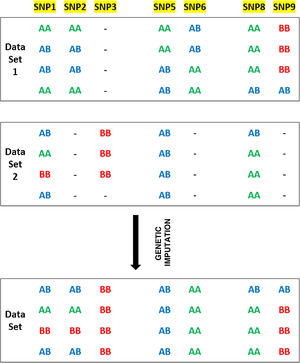
For example, one of the kits used by AncestryDNA has about 382,000 SNPs in common with another kit used by FamilyTreeDNA, meaning that approximately 300,000 SNPs do not coincide. Computing advances make it possible to use algorithms to complete or impute genotypes in markers that are not shared between two genotyping matrixes. Fig. 2 shows an example of genetic imputation.
SNPs 1-9 are located in three blocks with high binding imbalance. Data Set 1 and Data Set 2 represent the genotypes of eight individuals typified using different chips for SNPs 1-9. The final Data Set shows the expected results of genetic imputation. For example, although SNP2 is analysed in Data Set 1 it is not analysed in Data Set 2. Given the major binding imbalance between SNPs 1-3, the genotypes in Data Set 2 may be inferred on the basis of the genotypes present in Data Set 1 (https://www.illumina.com/content/dam/illumina-marketing/documents/products/technotes/technote_imputation_estimates_genotypes.pdf).
This method has an estimated error of from 2% to 6%, and this is chiefly due to two factors: the reference population (for example, some regions of the genome are more problematical in African populations, as they have fewer complete genotyped genomes of the same) and the software used (as different algorithms give rise to slight differences). It is therefore necessary to properly select the reference population we will use for the genetic imputation of our sample, and we also have to choose the software that fits the best with our needs.17
Arguments in favour and against the use of forensic genealogyIn favour- •
Its success speaks for itself (see the list of resolved cases in Appendix B supplementary file 2), as it has become the best investigation tool since the start of DNA-based identification.
- •
The individuals who commit violent crimes (who often reoffend) inflict great emotional and physical damage on their victims and their families. If a criminal career can be prevented as soon as it starts, future victims and the associated costs for them and society can be avoided. This would guarantee an ethical balance between individuals’ rights and privacy and independence, as opposed to public rights to personal safety and protection.6
- •
The United States criminal database contains (March 2020) more than 18 million criminal and detainee profiles18 which are regularly compared with almost 1 million crime scene profiles. Although this has enabled the resolution of about 450,000 criminal investigations in the last 20 years, many cases have yet to be resolved because the author cannot be identified. Only 40% of genetic profile searches in the database give results that make it possible to clear up a crime. Forty percent of homicides in the United States go unresolved, and more than 200,000 DNA kits corresponding to sexual crimes have not been analysed. Moreover, the criminal database does not cover all of the prison population of the United States.
- •
The white population is under-represented in criminal databases, although it is heavily over-represented in genealogical databases.19 The use of forensic genealogy may make it possible to balance the racial disparities in the American criminal justice system.20
- •
The methodology has not been validated for forensic usage21 and there is currently no training program or academic qualification for genetic genealogists. Insufficient information has been published on genotyping and identification methodology for a suitable peer review in scientific publications.
- •
The American Civil Liberties Union has expressed its concern about the use of public DNA databases for criminal investigations. It argues that when DNA profiles are published in public domains, information is also being published on their close or even distant relations, with potentially severe implications for the rights to privacy.22
- •
Some adopted people had believed that they had found their biological parents, only to subsequently discover that data had been wrongly interpreted. If this happens in cases of adoption searches it could also occur in a criminal investigation, with more serious emotional and reputational consequences (for example, as happened in an initial phase of the search for the “Golden State killer”, when a person was linked to this by searches in a Y chromosome database).21
- •
DNA evidence sometimes shows that an individual’s DNA has been found at a certain place, even though they had nothing to do with the crime in question: it may be on a soft drink can left at the crime scene, for example. Situations of this type could hardly be investigated in cases that are more than 40 or 50 years old, for example.
- •
Possible data loss or hacking of companies with genealogical databases.23 For example, a high percentage of the genotypes of other users with DNA data in a database could be extracted (up to 92% of markers with an exactitude of up to 98%).24
- •
False data could be entered so that certain individuals seemed to be related to other specific people, hindering possible lines of investigation.24
- •
It has been proven that it is possible to identify anonymised data (in scientific databases, for example)25 and this will continue to be a real threat26 until trustworthy and secure coding techniques are available. Data that is accessible in GEDmatch could be transferred to other online services (such as DNA.Land –htpps://dna.land– or Promethease –https://promethease.com) which offer data on susceptibilities to diseases. This could have an adverse effect on the possibilities for someone or their extended family to take out an insurance policy or to find work.
- •
There is no security if databases close or are acquired, or if they agree to collaborate with other companies: in July 2018 the GlaxoSmithKline [GSK] pharmaceutical company signed a four-year contract for 300 million dollars for the development of new drugs based on the 23andMe user database).
- •
The probability of being able to predict STR genotypes based on SNP haplotype data and vice versa.27 STR profiles could therefore be linked to SNP registries in public databases such as GEDmatch, and these could reveal not only identity information but also ancestor and health data.28
- •
Given that consanguinity and endogamy affect identifications, GEDmatch for example offers tools to check whether there parents of investigated individuals are related in any way, and this could reveal a history of incest in their family.
We have to distinguish between companies that commercialise DNA kits used for genealogy (FamilyTreeDNA, 23andMe, MyHeritage), companies which analyse the said kits (Bode Technology, Parabon Nanolabs) and investigation services or databases which store the results obtained (mainly GEDmatch and FamilyTreeDNA). Appendix B supplementary file 3 shows the characteristics of the most important forensic genealogy kit supply companies in terms of their genetic markers and users, etc.
Each company uses different coincidence thresholds for IBD detection, so that predictions may vary from company to company. They also offer different numbers of analysable SNPs, and some companies even offer complete genome sequencing.29
Mixes from two people can be analysed if the mix is dysbalanced and if the minority profile is available as a reference. It is even possible to analyse mixes from three people if the smallest contributor supplies mere traces. The material to be analysed may be blood, saliva, semen or bone, and the minimum amount of DNA necessary is 1ng.30
There are a large number of users:26 AncestryDNA (15 million users), FamilyTreeDNA (1 million users), MyHeritage (2.5 million users), 23andMe (10 million users), and GEDmatch (1.3 million users), considering that some individuals are users of two or more companies. It is estimated that the number of users in the databases of these companies will stand at about 100 million people by the end of 2021.31
GEDmatch is a website where users (the majority of whom are from the United States, followed by Canada, Australia and the United Kingdom) are able to upload the results of analyses carried out by FamilyTreeDNA, MyHeritage and most of the other genetic testing services. The higher the number of profiles in the database, the greater the probability of finding your family members. Although this site is free, by paying a monthly fee of 10 dollars it is also possible to use certain advanced search tools in the database.
The most widespread tools are those used to compare one’s own genetic profile with the whole database, to discover a list of individuals who share the largest amount of DNA segments with you, or to compare a certain individual in the database to see the chromosome locations and size of shared segments.
In the near future we will see a gradual implementation of forensic genealogy in criminal investigations by some of these companies. For example, after acquiring GEDmatch on 9 December 2019, Verogen, Inc. is now developing MPS kits for forensic use within the field of forensic genealogy.
Company policies respecting database access by police forcesUntil May 2019 police forces had no problem with access. Few cases were investigated and all of them concerned serious crimes (homicides and rapes). However, the police then used forensic genealogy tools to discover the author of a minor theft. This violated the GEDmatch policy, which restricted police usage to cases of homicide and sexual aggression. After the said investigation was made public, GEDmatch abruptly changed its policy: all users were now excluded by default (opted out) from police searches unless they had explicitly chosen to be included (opted in). After then, users had to approve a clause authorising sharing their data with police forces. After being acquired by Verogen, Inc., GEDmatch currently maintains this access policy. To date (January 2020), of the 1.3 million profiles in GEDmatch only 190,000 have accepted this clause.
On the other hand, from December 2018 FamilyTreeDNA users accept by default that the company permits searches for violent crime suspects (such as homicide or sexual violence) and the identification of cadavers (opting in). To date only 1.6% have specifically opted out, to maintain the privacy of their profiles. The company now has (on 31 January 2019) one million registries in its database. Company policy in Europe is different, as opt out is the default option there.
Other providers such as AncestryDNA (15 million users), 23andMe (10 million users) and MyHeritage (2.5 million users) attempt to prevent police forces from access the genetic data of their users. Nevertheless, these companies allow their users to download a digital copy of their DNA and load the corresponding file in public databases which are open to the police.
Of course, the police have access to all of the companies’ database registries if they have judicial authorisation to do so (depending on the crime and taking the proportionality of this measure into account).
International recommendations for the use of forensic genealogy in criminal investigationRecently, the United States Justice Department (11 January 2019)32, the American Society of Crime Laboratory Directors (18 October 2019)33 and the scientific work group on DNA analysis methods (SWGDAM, 18 February 2020)34 have published similar proposals for the criteria to be considered before undertaking analysis using forensic genealogy:
- •
Only serious crimes are to be analysed using forensic genealogy (homicide and sexual aggression), as well as for the identification of cadaveric remains.
- •
When traditional investigation has not supplied any clues and criminal database searches have not shown any coincidences.35
- •
When the DNA to be analysed can only be attributed to the person of interest in each case in question.
- •
Laboratory staff, genealogical investigators, judges and public prosecutors must all be trained and qualified in the use of this technology.
- •
The police will have to gain judicial authorisation to process forensic genealogy cases.
- •
The privacy of results must be guaranteed together with the transparency of the methods used, to ensure public trust in the system is maintained.
- •
Once an individual has been identified using forensic genealogy, the police will have to obtain a reference sample from the same for conventional analysis of STR markers for the purpose of exclusion/inclusion.
- •
If inclusion arises from the said analysis then the profile entered will be eliminated from the genealogical database.
It would also be recommendable for the International Society of Forensic Genetics (ISFG) to publish recommendations for the use of this tool in forensic investigations. The ISFG itself is already aware of the importance of this tool, and thus the Full Meeting of the last congress (Prague, September 2019), approved the creation of a new workgroup on forensic genealogy led by the famous investigator Chris Phillips (Santiago de Compostela University).
Other recommendations for the correct use of genealogical databases24,36- •
Only data which have been verified37 by the use of cryptographic techniques should be used,38 which are able to trace a genetic data file to a company, genotyping chip and specific date.
- •
The integrity of genetic profiles must be guaranteed by anomaly detection, to prohibit the inclusion of irregular data in genealogical databases.
- •
Only comparisons with genetic profiles which show high quality coincidences should be allowed.
- •
The use of tools that permit high resolution chromosome visualisations and obtaining precise coordinates for coinciding segments should be restricted.
This review shows the usefulness of forensic genealogy as an investigation tool in resolving crimes or identifying cadaveric remains. Nevertheless, we have to ensure that a balance is kept between personal privacy and the interests of the individuals affected, on the one hand, and public safety, the common good and the resolution and clearing up of crimes on the other.
Consumers in Europe enjoy a higher level of legal security with products of this type in terms of the safeguard of their personal data and exchange of information between third parties,39 independently of whether individual countries have stricter regulations in this field. In fact, in Europe 23andMe, for example, applies the opt-out option by default for the authorisation of searches in connection with criminal investigations in its database.
In Spain there is, additionally, the existence of Organic Law 10/2007, of 8 October, which governs the police database of DNA-based identifiers and does not cover the use of forensic genealogy. However, certain aspects of this law were outdated by the time it was passed, as the lawmakers could not foresee the speed of genomic discoveries. We therefore find gaps in the law, not only in this field, but also regarding the use of codifying markers, searches for family members in the police database and the use of ancestry-informative markers and phenotypes, etc.
The aim of this work is to offer information and, as far as is possible, to generate a debate which will lead to suitable regulation of these practices. This debate has already started, at least in scientific terms, within the National Commission for the Forensic Use of DNA in Spain. Nevertheless, it has to expand to include academic sectors of Legal Medicine, the world of the judiciary, the field of Bioethics and society in general. We hope that this paper furthers this aim.
Conflict of interestsThe author has no conflict of interests to declare.
Please cite this article as: García Ó. Genealogía forense. Implicaciones sociales, éticas, legales y científicas. Rev Esp Med Legal. 2021;47:112–119.