Multiple factors, including both genetic and environmental mechanisms, appear to play a role in the aetiology of headache. An interesting area of study is the possible involvement of epigenetic mechanisms in headache development and the transformation to chronic headache, and the potential role of these factors as a therapeutic target.
MethodsWe performed a literature review of the involvement of different epigenetic mechanisms in headache, mainly using the Medline/PubMed database. To this end, we used the following English search terms: headache, migraine, epigenetics, DNA methylation, histones, non-coding RNA, and miRNA.
ResultsA total of 15 English-language publications related to the above terms were obtained.
ConclusionThere is limited but consistent evidence of the relationship between epigenetics and headache; it is therefore essential to continue research of epigenetic changes in headache. This may help to understand the pathophysiology of headache and even to identify candidate biomarkers and new, more effective, therapeutic targets.
En las cefaleas parece existir una influencia multifactorial, tanto de mecanismos genéticos como ambientales, siendo interesante el estudio de la posible participación de mecanismos epigenéticos en su desarrollo, cronificación y potencial papel como diana terapéutica.
MétodosHemos llevado a cabo una revisión bibliográfica, principalmente a través de la base de datos Medline/PubMed, de la implicación de los distintos mecanismos epigenéticos en las cefaleas. Para ello hemos utilizado los términos de búsqueda en inglés: headache, migraine, epigenetics, DNA methylation, histones, non-coding RNA y miRNA.
ResultadosSe obtuvieron un total de 15 publicaciones en idioma inglés relacionadas con los términos anteriores.
ConclusionesExisten indicios de la relación entre la epigenética y las cefaleas, siendo imprescindible, debido al reducido número de estudios, continuar con la investigación de las modificaciones epigenéticas en las cefaleas. Esto podría ayudar a comprender la fisiopatología de las cefaleas e incluso identificar biomarcadores y nuevas dianas terapéuticas más eficaces.
Epigenetics is the study of the mechanisms that regulate gene expression without modification of the genetic code. These mechanisms play an essential role in cell and tissue differentiation during embryogenesis and in long-term response to environmental stimuli in adult organisms. The main epigenetic modifications are DNA methylation, gene silencing by non-coding RNA, and histone post-translational modifications. DNA methylation in eukaryotes consists in the addition of a methyl group to the carbon 5 position of the cytosine ring via DNA methyltransferase, modifying DNA function when it occurs at promoter regions, and frequently repressing gene transcription. Modifications to histone N-terminal tails, such as histone methylation or acetylation, determine chromatin structure and chromatin's tendency to gene transcription (euchromatin) or to gene expression silencing (heterochromatin). Non-coding RNAs are RNA molecules of variable size and function that do not code for proteins; one example are micro-RNAs (miRNA). Their role in the regulation of gene expression is currently under study. miRNAs can bind to messenger RNAs (mRNA), regulating post-transcriptional gene expression through interference with the translation of the mRNA into a protein.
Relevance of epigenetic mechanisms of migraine in clinical practiceAwareness has increased in recent years of the role of epigenetic mechanisms on a wide range of multifactorial disorders. One such condition is headache, a highly prevalent and disabling nervous system disorder whose pathogenesis involves numerous genetic and environmental mechanisms. In the light of this, it seems reasonable to hypothesise that the pathophysiology and chronification of headache involve epigenetic mechanisms. For example, environmental factors may trigger or at least increase the risk of developing migraine attacks, and probably contribute to migraine to the same extent as do genetic factors.1 Epigenetic mechanisms contribute to the development of disease phenotypes by mediating the effect of environmental factors on gene expression. Current symptomatic and preventive treatments for headache are not completely effective; migraine treatment, for instance, is effective in less than half of patients.2 Determining predisposing factors, including epigenetic factors, is therefore essential to identifying new molecular targets, with a view to developing more effective treatments for headache. Furthermore, patient response to analgesics and triptans may change over time, depending on the frequency of use. This cannot be explained exclusively by alterations in the genetic code; rather, it suggests the involvement of epigenetic changes in target molecules. We review the literature on the association between epigenetic mechanisms and headache; the study was conducted in July 2017. We searched the Medline/PubMed database using the following keywords: headache, migraine, epigenetics, DNA methylation, histones, non-coding RNA, and miRNA. The literature search identified 15 articles, all of which were written in English.
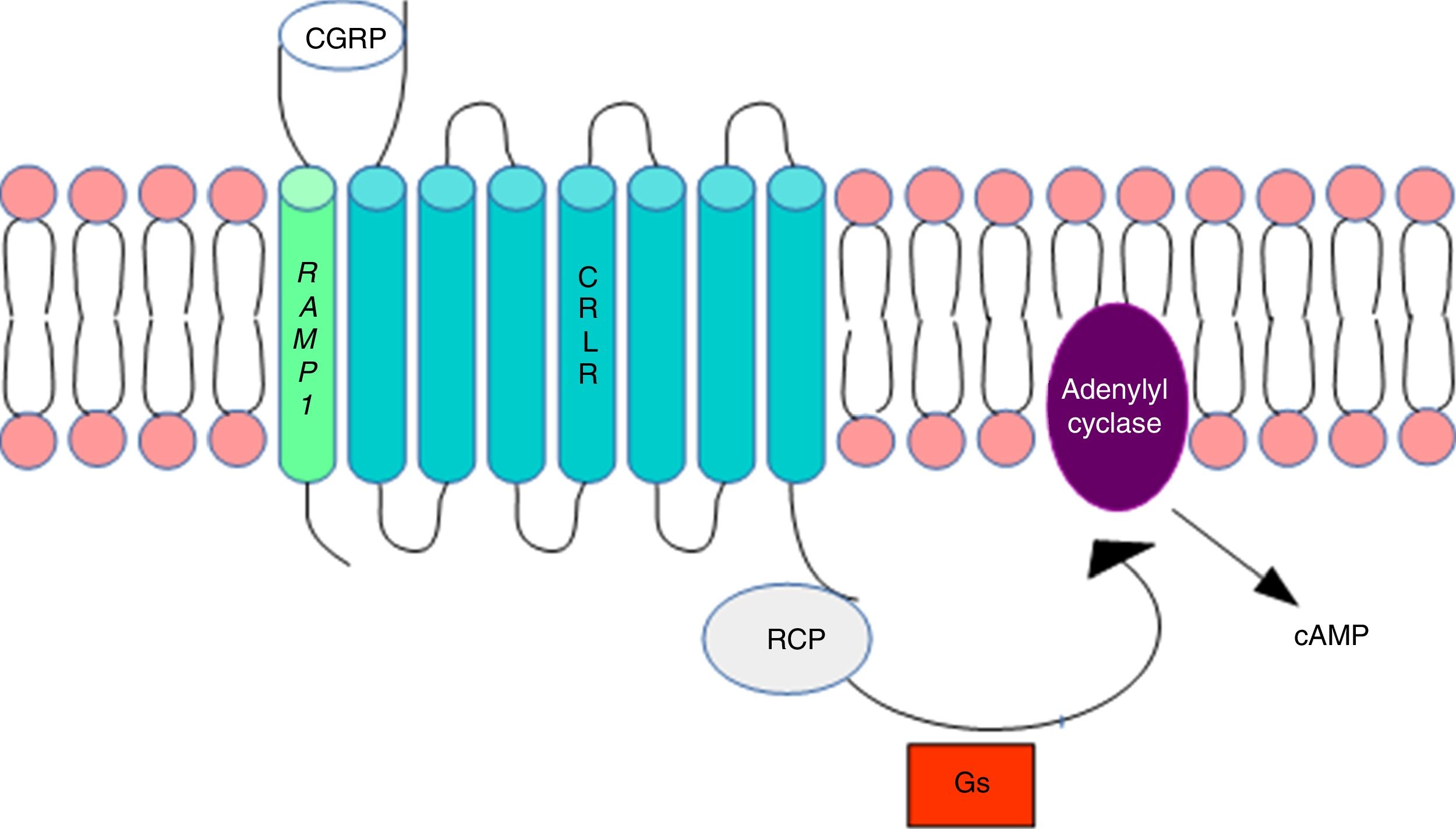
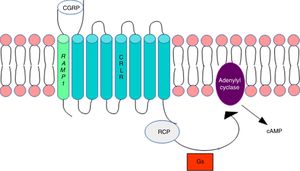
DNA methylation in headacheThe calcitonin gene pathwayThe CALCA gene codes for calcitonin gene-related peptide (CGRP); by alternative splicing, it also codes for calcitonin. CALCA expression is normally limited to endocrine cells and neurons; the gene is not expressed by glial cells. The RAMP1 gene, which together with the calcitonin receptor-like receptor (CRLR) forms the CGRP receptor, is involved in the regulation of CGRP activity (Fig. 1).
Calcitonin gene-related peptide receptor. Formation of the calcitonin gene-related peptide in the cell membrane. Modified from Ramos-Romero and Sobrino-Mejía.51
CGRP: calcitonin gene-related peptide; CRLR: calcitonin receptor-like receptor; G: G protein; RCP: receptor component protein.
Elevated CGRP levels have been associated with migraine pathogenesis. Several studies provide evidence of CGRP's involvement in the pathophysiology of migraine: CGRP injection has been found to induce migraine-type headache in patients with migraine but not in healthy controls,3 and a CGRP receptor antagonist is reported to have analgesic effects.4–6 This may suggest that regulation of the CALCA and RAMP1 genes is involved in migraine susceptibility.
Wan et al.7 analysed the association between migraine and the DNA methylation pattern at the RAMP1 promoter region in peripheral leukocytes. Fifty-one blood DNA samples (from 26 patients with migraine and 25 healthy controls) were treated with bisulfite and DNA methylation levels were subsequently measured at the RAMP1 promoter region. The researchers found no significant differences between patients and controls in methylation levels in 13 CpG sites (in the gene promoter region, at positions −300 to +205bp from the transcription start site), but they did observe a mild trend towards lower methylation in patients with migraine. Interestingly, subsequent stratification analysis revealed that the methylation level at the +25, +27, +31 CpG unit was significantly associated with family history of migraine, and methylation at the +89, +94, +96 CpG unit was correlated with migraine in women. Curiously enough, when the methylation level at the +89, +94, +96 CpG unit dropped below 3.50%, the risk of migraine increased significantly in women, but not in men. The authors suggest that the methylation level in this region may constitute an epigenetic biomarker of migraine risk in women. Despite its limitations (small sample, use of leukocytes), the study provides evidence that DNA methylation at the RAMP1 promoter region may play a role in migraine pathogenesis.
In view of mounting evidence of the involvement of glial cells in pain, Park et al.8 analysed whether cell-specific CALCA gene expression is caused by epigenetic mechanisms. The authors used rat and human model cell lines and primary cultures of rat trigeminal ganglia glia to measure DNA methylation and histone acetylation at a CpG island near a promoter region (the 18-bp enhancer), which is active in neurons, though inactive in glial cells. They found that DNA methylation and histone H3 acetylation at the CpG island were correlated with CALCA gene expression; hypomethylation was observed in cells expressing the gene, and hypermethylation and hypoacetylation in non-expressing cells (glia). The cell-specific patterns of histone methylation and acetylation may therefore explain both rat CALCA and human CALCA gene silencing in different cell types. Park et al.8 also analysed the functional consequences of altering the chromatin state, taking quantitative measurements of calcitonin and CGRP mRNAs after using the methylation inhibitor 5-aza-2′-deoxycytidine and the histone deacetylase inhibitor trichostatin A. DNA treatment with 5-aza-2′-deoxycytidine induced CALCA expression both in human and in rat cells and in glial cultures. Although trichostatin A alone had no effect, trichostatin A plus 5-aza-2′-deoxycytidine had a strong synergistic effect on CALCA gene induction in glia. This suggests that DNA methylation is necessary for histone acetylation, with methylation in this region being essential for cell-specific CALCA gene expression. In conclusion, the study by Park et al.8 shows that epigenetic modulation is sufficient to induce CALCA expression in glial cells. Given that the CALCA gene is systemically activated by inflammation in non-CALCA-expressing tissue, mainly producing procalcitonin (a precursor to calcitonin), and the suggestion that neurogenic inflammation is involved in migraine pathogenesis, procalcitonin may constitute a biomarker of migraine.8
In view of the difficulty of obtaining human nerve tissue, the studies cited previously used human leukocytes, rat cell lines, or rat nerve tissue. However, it is unclear whether DNA methylation patterns in blood leukocytes are correlated with those observed in nerve tissue. Labruijere et al.9 used rats to compare methylation in different genes that are probably epigenetically regulated and are involved in migraine pathogenesis, including CALCA, RAMP1, CRCP, and CALCRL, in leukocytes and tissues involved in migraine pathophysiology (dura mater, trigeminal ganglion, caudal trigeminal nucleus). A correlation was observed in methylation patterns in the CALCA and RAMP1 genes in all tissues; the same was not true for other genes. The results of previous studies of CALCA and RAMP1 may therefore be extrapolated to nervous tissue. Labruijere et al.9 also report a strong correlation between DNA methylation in rat and human leukocytes, and suggest that DNA methylation may be studied in rat tissue when human tissues are difficult to obtain.
Headache chronificationThe biological mechanisms involved in transition from episodic to chronic headache are yet to be discovered. However, given the capacity of changes in DNA methylation to alter the state of biological systems, the involvement of these changes in headache chronification is a promising field of study.
The involvement of DNA methylation mechanisms in headache chronification is based on the fact that synchronous neuronal activity (also present in cortical spreading depression [CSD] in migraine with aura) changes DNA methylation of genes associated with neuronal plasticity.10
A study by Winsvold et al.11 supports the hypothesis that methylation changes in genes regulating synaptic plasticity are associated with headache chronification. The study included 36 women presenting headache chronification between baseline and after 11 years of follow-up, and 35 female controls with episodic headache. DNA methylation was subsequently quantified at 485000 CpG sites; differences in methylation between patients and controls were analysed in 2 stages by linear regression. The researchers identified 20 CpG sites that may signal genes involved in headache chronification, although none reached statistical significance in a fixed-effects meta-analysis (1.15×10−7). The CpG sites with the strongest association with headache chronification were located in SH2D5 and NPTX2, both of which regulate synaptic plasticity. When considering the more extensive list of the 200 CpG sites most strongly associated with headache chronification, statistically significant enrichment was observed for genes involved in calcium ion binding. However, the study's cross-sectional design prevents us from determining whether methylation changes in these genes are a cause or rather a consequence of frequent headache episodes.
Histone modification in headacheHistone methylation is a process by which one, 2, or 3 methyl groups are transferred to some amino acids in histone tails. Each level of modification may have different biological effects depending on the context and the residues affected. Histone regulation, and consequently chromatin remodelling, plays a critical role in regulating gene transcription and promoting long-term changes in neuronal plasticity. In addition to the hypothetical involvement of histone H3 acetylation in CALCA expression,8 other changes in histone acetylation are reported to be associated with headache.
Histone modifications in cortical spreading depressionCSD is thought to be a pathophysiological mechanism activating the trigeminovascular system. This phenomenon is characterised by a wave of depolarisation of neurons and glial cells that propagates slowly across the cerebral cortex, followed by sustained suppression of spontaneous neuronal activity. Cortical changes and metalloproteinase activation occurring during CSD disrupt the blood-brain barrier, allowing chemical mediators to activate the trigeminal nerve terminals surrounding meningeal blood vessels.12 CSD also seems to participate in epigenetic control of gene expression by inducing histone modifications. Trimethylation of lysine 4 at histone H3 (H3K4) occurs in all active genes, whereas H3K9 trimethylation occurs in transcriptionally inert compact heterochromatin.
Passaro et al.13 analysed whether CSD causes epigenetic modifications in rat chromatin by comparing the methylation levels of H3K4 and H3K9 between the brain hemisphere where CSD occurs and the contralateral hemisphere, 24hours after inducing CSD. The researchers observed epigenetic chromatin modifications 24hours after CSD induction, with significant decreases in H3K4 dimethylation and monomethylation and increased H3K9 dimethylation. Rana et al.14 used rats to study these changes in 2 neuroprotective genes, iNOS and HIF1ɑ, confirming the hypothesis that CSD can affect epigenetic regulation of gene expression.
Polymorphisms in “epigenetic” genes associated with medication overuse in patients with headacheHeadache medication overuse may share pathogenic mechanisms with other types of drug addiction; these may involve genetic factors predisposing individuals to medication overuse.15–17 Histone deacetylase 3 inhibition has been found to play a role in the memory processes involved in extinction of drug-seeking behaviour in animal models.18 This protein, which is expressed in nearly all tissues, including the brain, is responsible for deacetylation of lysine residues in core histones. Pisanu et al.19 were the first to study the role of histone deacetylase 3 polymorphisms in patients with medication overuse. Despite a small sample size, the study revealed a significant association between the G allele of the rs2530223 polymorphism and greater medication use, although no significant correlations were observed with headache frequency or intensity.
Involvement of the JNK signalling pathway in epigenetic regulationMigraine patients present marked hyperexcitability of the brain, which reacts atypically to external and internal stimuli, potentially triggering a migraine attack. Trigeminal nerve fibres in the oral and nasal cavities are sensitive to multiple environmental stimuli, such as chemical irritants and toxins. JNK-mediated phosphorylation of inducible transcription factor c-Jun greatly determines its potential to activate gene transcription. Phosphorylation represents the last step of a cascade of signals in the response to nerve fibre lesions and nervous system infection and inflammation.20–23 Wu et al.24 conducted an in vitro study to evaluate the role of the JNK/c-Jun cascade in the regulation of histone H3 acetylation in rat trigeminal neurons following exposure to a neurotoxic stimulus (mustard oil), as well as the level of histone acetylation in response to this stimulus. This was the first study to provide solid evidence of JNK's involvement in the epigenetic modulation of histones in peripheral trigeminal neurons in response to chemical environmental stimuli.24
Changes in non-coding RNAInstead of being translated into proteins, non-coding RNA molecules regulate gene expression. Among the broad range of non-coding RNA molecules, miRNAs have been studied extensively in recent years. miRNAs reduce mRNA levels and play an important role in post-transcriptional gene regulation by forming RNA-induced silencing complexes. miRNAs also appear to be involved in pain signalling: miRNA dysregulation has been observed in patients with complex regional pain syndrome, osteoarthritis, and fibromyalgia.25–27 This has led researchers to suggest that miRNA is altered in migraine, although only a limited number of preliminary studies have been conducted.
Andersen et al.28 examined serum microRNA alterations in patients with migraine and in healthy controls and evaluated whether migraine manifests in chronic serum miRNA alterations. High serum levels of miRNA-34a-5p and miRNA-382-5p were observed both during migraine attacks and during pain-free periods. Interestingly, miRNA-34a-5p has been associated with inflammation and vascular endothelial stress response, whereas miRNA-382-5p is observed mainly in neurons and cerebrospinal fluid, and is present only in small amounts in serum. The blood-brain barrier is thought to be disrupted during migraine attacks, which suggests that increased miRNA-382-5p levels may originate in central nervous system structures or in the cerebrospinal fluid. The authors conclude that migraine alters serum miRNA expression not only during attacks, but also during pain-free periods, which points to the important role of miRNAs in the pathophysiology of migraine. According to these results, serum miRNA levels may serve as biomarkers of migraine for potential applications in stratification, diagnosis, and treatment monitoring.
Tafuri et al.29 evaluated circulating miRNA expression in women with migraine without aura during pain-free periods and in healthy controls. The researchers observed a specific profile of circulating miRNA associated with migraine: significant miRNA-27b up-regulation and miRNA-181a, miRNA-let-7b, and miRNA-22 down-regulation as compared to controls. Furthermore, the specificity and sensitivity of the miRNA profile for diagnosing migraine were comparable to those of the gold-standard clinical criteria; evaluating these 4 miRNAs may constitute a powerful diagnostic tool for migraine without aura.
Neuroimmunological alterations of the central and peripheral nervous systems play an essential role in the pathophysiology of chronic pain30–36; non-coding RNAs,37 and particularly miRNAs, regulate both immune and neurological processes. New evidence suggests that endogenous analgesic systems, including the GABAergic and opioid systems, are regulated by miRNAs including miRNA-134 and miRNA-181a.38,39 Furthermore, specific miRNAs have been associated with neuropathic pain and dysregulation of ion channel gene expression in mouse models.38,40,41 Similarly, understanding of the dysregulation of neuronal miRNAs may be applied not only to neuropathic pain but also to such other painful syndromes as headache, particularly hereditary migraine and other types of migraine.
Dopaminergic and glutamatergic signals from the amygdala, hippocampus, and prefrontal cortex to the nucleus accumbens are involved in impulse control circuits, whereas the nucleus accumbens is involved in emotional aspects of pain processing. An association between chronic pain and emotional dysfunction has been observed; maladaptive responses from the nucleus accumbens in neuropathic pain have been associated with miRNA dysregulation in this region.42 This suggests that pain may alter the expression of certain miRNAs in primary nociceptors and brain regions associated with the emotional component of pain.42–50
In conclusion, research on miRNA profiles provides information on potential biomarkers for predicting the onset, severity, and progression of migraine. miRNAs may also serve as therapeutic targets, leading to radical changes in migraine management.
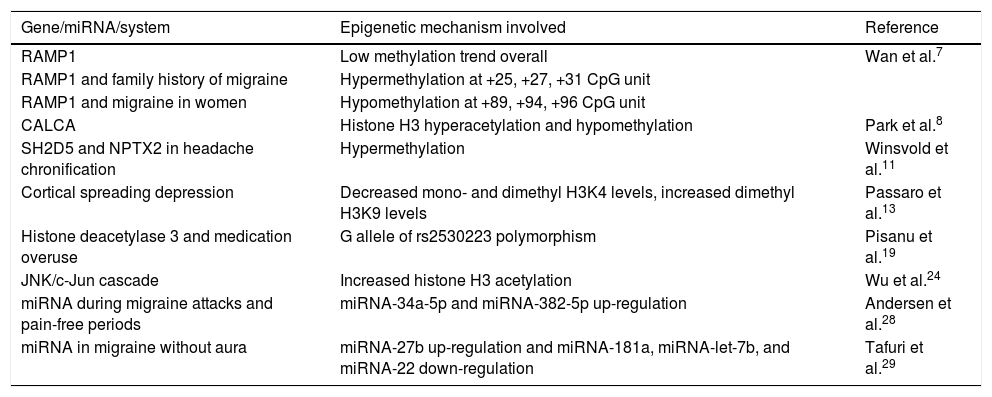
Table 1 summarises the genes, miRNAs, and epigenetically modified mechanisms of migraine identified in our literature review.
Epigenetically modified genes and mechanisms involved in headache.
| Gene/miRNA/system | Epigenetic mechanism involved | Reference |
|---|---|---|
| RAMP1 | Low methylation trend overall | Wan et al.7 |
| RAMP1 and family history of migraine | Hypermethylation at +25, +27, +31 CpG unit | |
| RAMP1 and migraine in women | Hypomethylation at +89, +94, +96 CpG unit | |
| CALCA | Histone H3 hyperacetylation and hypomethylation | Park et al.8 |
| SH2D5 and NPTX2 in headache chronification | Hypermethylation | Winsvold et al.11 |
| Cortical spreading depression | Decreased mono- and dimethyl H3K4 levels, increased dimethyl H3K9 levels | Passaro et al.13 |
| Histone deacetylase 3 and medication overuse | G allele of rs2530223 polymorphism | Pisanu et al.19 |
| JNK/c-Jun cascade | Increased histone H3 acetylation | Wu et al.24 |
| miRNA during migraine attacks and pain-free periods | miRNA-34a-5p and miRNA-382-5p up-regulation | Andersen et al.28 |
| miRNA in migraine without aura | miRNA-27b up-regulation and miRNA-181a, miRNA-let-7b, and miRNA-22 down-regulation | Tafuri et al.29 |
Headache, and particularly migraine, has a considerable impact on health and society. The aetiopathogenic mechanism of headache is yet to be fully understood, and the symptomatic treatments currently available are not sufficiently effective; further research on the subject is therefore essential. Given that headache pathogenesis is thought to involve both external and genetic mechanisms, epigenetics provides an interesting perspective as it represents the interaction between both types of factors.
Despite the small number of studies on the topic, preliminary data point to an association between epigenetic mechanisms and headache. It is therefore essential to continue this line of research to identify the epigenetic mechanisms involved in the pathophysiology of migraine, determine their usefulness as potential biomarkers, and identify molecules modulating chromatin structure in migraine, which may be therapeutic targets for migraine treatment.
FundingThe study has received no funding of any kind.
Conflicts of interestThe authors have no conflicts of interest to declare.
Please cite this article as: Cámara MS, Martín Bujanda M, Mendioroz Iriarte M. Modificaciones epigenéticas en las cefaleas. Neurología. 2021;36:369–376.
This study has not been presented at the SEN's Annual Meeting or at any other meetings or congresses.