In the present manuscript we describe the planning carried out in our hospital to adapt our diagnostic capability to perform large numbers of SARS-CoV-2 RT-PCR.
MethodsThe analysis and prediction of workflow included the number of RT-PCR per week from the beginning of the pandemic, with a total of 31971 determinations. The planning phase was developed based on the different scenarios previously predicted.
ResultsAccording to the predictions obtained, an automated custom solution was chosen, based on the use of the OT-2 open-source liquid-handling robots (Opentrons), to design a reproducible workflow that achieved a production capacity of 5640 samples/day, with a time of response of four hours per procedure.
ConclusionsThe analysis and prediction of workflow, along with the use of the robotic platforms OT-2, provided a robust structure to deal with the high demand of determinations that this pandemic requires.
Este artículo describe la planificación efectuada para adecuar la capacidad diagnóstica de grandes volúmenes de RT-PCR de SARS-CoV-2.
MétodosEl análisis y predicción del flujo de trabajo incluyó el número de RT-PCR desde el inicio de la pandemia, con 31971 registros. La planificación de la capacidad y de las opciones diagnósticas se planteó en base a los posibles escenarios derivados de las predicciones efectuadas.
ResultadosDe acuerdo con las predicciones obtenidas, se optó por una solución automatizada basada en el empleo de robots OT-2 (Opentrons) para configurar un flujo de trabajo reproducible, que logró una capacidad de procesamiento de 5.640 muestras/día, con un tiempo de respuesta de cuatro horas.
ConclusionesEl análisis y predicción del flujo de trabajo, unido al empleo de plataformas basadas en OT-2, proporciona una infraestructura robusta que permite atender con éxito las demandas de prueba que exige esta pandemia.
During the first wave of the pandemic, diagnostic capacity in hospitals using Reverse Transcription Polymerase Chain Reaction (RT-PCR) for SARS-CoV-2 was insufficient to cope with the huge demand. This was partly because the situation changed from RT-PCR being used as a clinical diagnostic tool to it being used as an epidemiological diagnostic tool.
At the beginning of September 2020, before the publication of the Sociedad Española de Enfermedades Infecciosas y Microbiología Clínica’s (SEIMC) [Spanish Society of Infectious Diseases and Clinical Microbiology] recommendations on the organisation of the diagnosis of SARS-CoV-21, we detected an increase in the number of samples for diagnosis by SARS-CoV-2 RT-PCR which, based on the Centers for Disease Control criteria2, coincided with a possible progression from scenario 2 (viral circulation with localised outbreaks) to scenario 3 (community transmission) in our health area. At that point, we began to optimise diagnostic decision-making by planning for possible evolving scenarios.
Methods and practicalitiesAnalysis and prediction of workflowThe analysis included the number of weekly RT-PCR tests since the beginning of the pandemic and included 31,971 records. Considering the presence of a trend in this period, we calculated a prediction model using the Holt3 double exponential smoothing technique to forecast the number of samples expected for weeks 37–44 (September and October). We selected the model using the minimum Bayesian Information Criterion (BIC) and confirmation of the absence of residual autocorrelation using the Ljung-Box test4. The analysis was performed with the IBM SPSS v.21 (IBM, Armonk) statistical program.
Planning of the need for a progressive escalation of diagnostic capacityThe planning of the capacity and the available diagnostic options was proposed according to the possible scenarios based on the predictions made and the recommendations of the Innovative Genomics Institute SARS-CoV-2 Testing Consortium5.
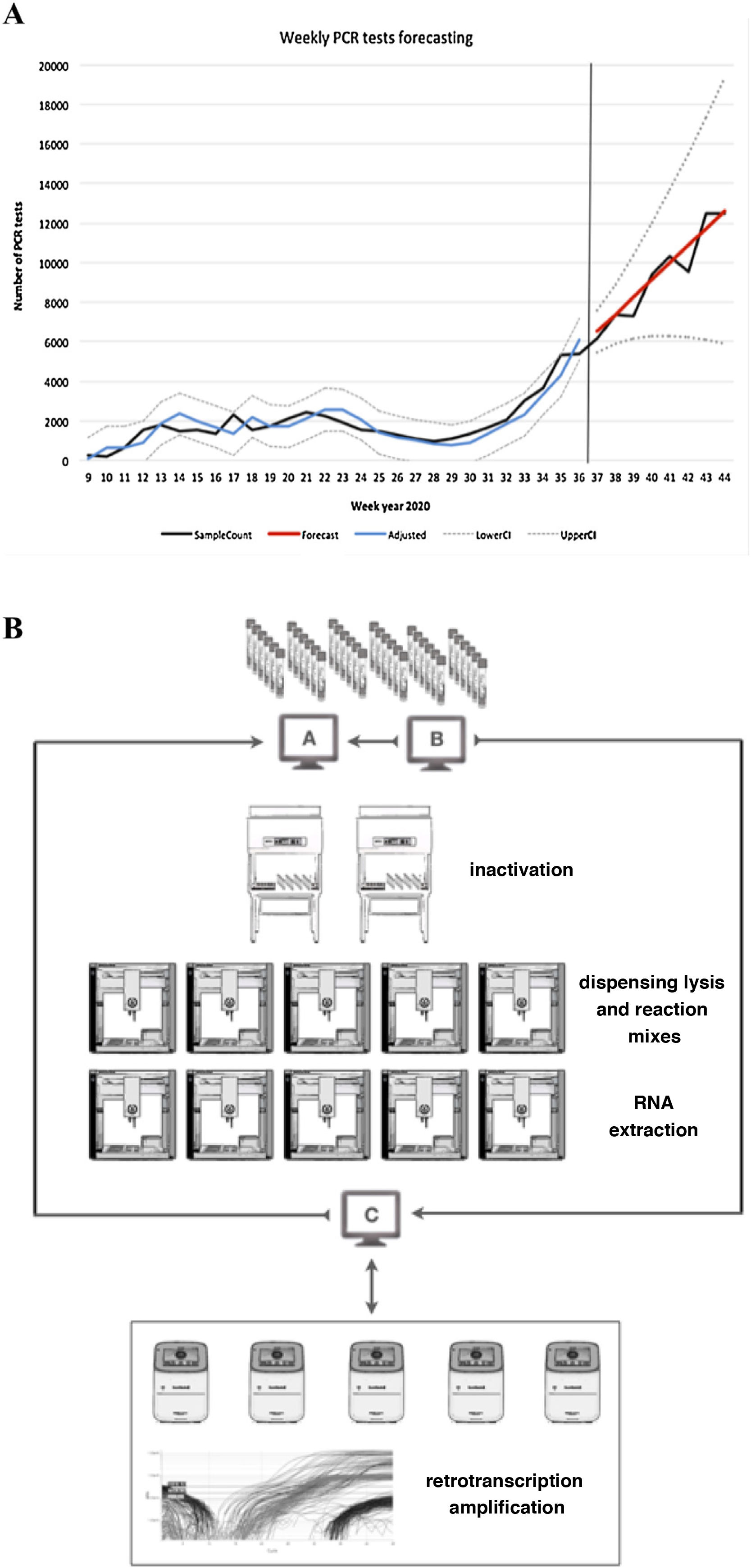
ResultsAnalysis and prediction of workflowThe prediction suggested three possible scenarios according to the mean values and confidence intervals (optimistic, likely and pessimistic). The values predicted by the model, together with their 95% CI, are shown in Table 1 and Fig. 1A.
Values forecast by the workflow prediction model for performing SARS-CoV-2 RT-PCR.
| Week | Number of RT-PCR | Forecast (likely scenario) | Lower confidence interval (optimistic scenario) | Upper confidence interval (pessimistic scenario) | Observed-predicted differences |
|---|---|---|---|---|---|
| 37 | 6,154 | 6,519 | 5,453 | 7,586 | −365 |
| 38 | 7,378 | 7,392 | 5,875 | 8,909 | −14 |
| 39 | 7,307 | 8,264 | 6,128 | 10,400 | -957 |
| 40 | 9,448 | 9,136 | 6,259 | 12,014 | 312 |
| 41 | 10,325 | 10,009 | 6,291 | 13,726 | 316 |
| 42 | 9,548 | 10,881 | 6,240 | 15,521 | −1,333 |
| 43 | 12,468 | 11,753 | 6,115 | 17,391 | 715 |
| 44 | 12,478 | 12,625 | 5,921 | 19,330 | −147 |
A) Prediction of workflow. SampleCount: actual samples, Forecast: prediction (likely prediction).
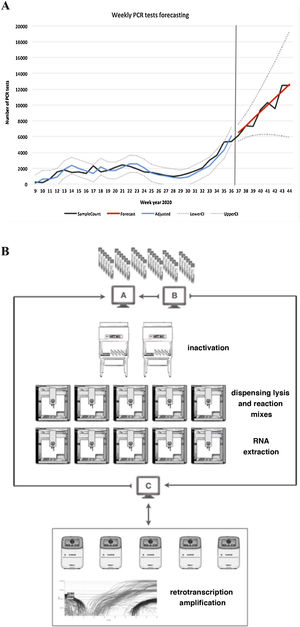
Adjusted: adjusted model, LowerCI: lower confidence interval (optimistic forecast), UpperCI: upper confidence interval (pessimistic forecast). B) Sample workflow based on OT-2 modules.
A: laboratory computer system, B: traceability program, C: programming-validation program.
From the predictions obtained, we chose an automated and scalable solution based on the use of open-source OT-2 liquid handling robots (Opentrons) to configure a reproducible workflow for SARS-CoV-2 RT-PCR testing. This design permitted the optimisation of the number of robots used per line of work (Fig. 1B).
In the analytical phase, the workflow was automated and standardised using external software which guaranteed traceability at all times. The design of the analytical phase involved the use of manual and robotic processes according to the terms of time performance.
The inactivation process was carried out manually in a biosafety cabinet external to the OT-2 due to the great variety of sample vials received.
The automated process involved the commissioning of five working lanes with two OT-2 modules per line. The OT-2 were configured in asynchronous mode and were used progressively based on the demand for tests. In each lane, the first OT-2 (station A) was configured to automate two independent processes: the dispensing of the lysis mixture into the sample wells and the dispensing of both the reagents necessary for the extraction process and the reagent mixture. The second OT-2 (station B) was configured to perform the RNA extraction process and the preparation of the 96-well reaction plates. Reverse transcription and amplification processes were performed externally in QuantStudio 5 thermocyclers (Thermofisher Scientific). In this process, an intermediate program (in-house application programmed in VBA) connected to the traceability program and made it possible to program the same positions in the thermocycler software.
In the post-analytical phase, the results generated by the thermocyclers were exported back to the intermediate program and then transformed into a user-friendly table, which included an automatic interpretation of the results.
System performanceIn terms of throughput, once the working times overlapped, 94 samples could be processed and validated every two hours per OT-2 lane, leading to a maximum throughput per lane of approximately 1,128 samples/day. The 5 OT-2 system (5 × station A, 5 × station B) working in parallel would yield a maximum throughput of 5,640 samples/day, above the estimate provided by the model.
This working protocol made it possible to process and validate the large volume of samples received from the health area every day in very little time. According to the method established for reporting results to the health area, 21.6% were reported within eight hours of being received by the laboratory, and 90% within 24 h.
DiscussionFrom the analytical standpoint, the pandemic has brought two major problems to light: the diagnostic capacity of laboratories and another very significant logistics issue6,7. To respond to this demand, most laboratories have relied on “closed” commercial platforms. These systems respond poorly both to large increases in demand, as they are not scalable, and to the need to include new tests. Additionally, in the event of a significant increase in demand, these systems can have logistics problems if there is a shortage in any part of the manufacturer’s reagent production supply chain8. However, open-source platforms such as OT-2 deliver big advantages over commercial platforms9,10, particularly in their flexibility and rapid adaptation to diagnostic needs and adaptation to available reagents, as well as in terms of profitability11.
Nonetheless, establishing a reproducible workflow based on OT-2 involves a number of critical points. From the organisational standpoint, there is the need to integrate processes that are external to the OT-2. Sample inactivation is a step that could be easily integrated into the OT-2, although it proved impossible due to the variety of types of transport vials received. Another critical point is integration with the amplification systems, a problem we solved by using traceability software, which allowed the automatic programming of the thermocyclers. Analytically, a critical point is the extraction of RNA12, an aspect that can make the difference between using OT-2 as liquid dispensing systems and turning them into molecular diagnostic platforms. The adaptation of magnetic modules to the OT-2 made it possible to include this process in the workflow and to reduce the use of external equipment. Another critical point occurs in the post-analytical phase, with the review and validation of a large volume of results, which in our case was resolved by implementing a computer solution that facilitated and automated the process.
Despite the improvement in processing large volumes of samples, and as is indicated in the results, we did not manage to report 100% of the samples within 24 h. The reason was that the samples were arriving in large volumes (in batches of more than 1,000), meaning that at times all five lanes were busy and samples were left awaiting processing. In addition, all the samples had to be correctly registered before they were processed to ensure traceability.
Despite the foregoing, the workflow with the OT-2 enabled us to adapt to the expected scenario. By way of example, comparing our solution based on OT-2 modules to other commercial platforms, response times are slightly higher, 4 h compared to 3.5 h for the Cobas SARS-CoV-2 RT-PCR (Roche Diagnostics) or 2.5 h for the Panther Fusion SARS-CoV-2 RT-PCR (Hologic, Inc) (although it requires prior inactivation). However, diagnostic capacity is much more versatile; working at full capacity, the OT-2 modules enable 5,640 samples to be processed per day, compared to 1,440 and 1,150 with the Cobas and Panther platforms, respectively13, unless several platforms are used together.
Our design supports the use of open PCR platforms, which are currently handling a significant proportion of the SARS-CoV-2 RT-PCR tests being processed in a number of hospitals and research centres14.
The authors thank the NGO COVIDWarriors for donating the Opentrons OT-2 stations to Hospital Universitario Virgen del Rocío (Seville, Spain) and Vitro S.A and Roche Diagnostics for their help in the implementation of the platform.
Please cite this article as: Camacho-Martínez P, Martin-Gutiérrez G, Peñalva G, Merino-Díaz L, Lepe JA. Optimización y adecuación de la capacidad diagnóstica para la realización de grandes volúmenes de RT-PCR de SARS-CoV-2. Enferm Infecc Microbiol Clin. 2022;40:441–444.