Edited by: Sonia Roman
Last update: January 2023
More infoObesity is a global health problem that triggers fat liver accumulation. The prevalence of obesity and the risk of non-alcoholic steatohepatitis (NASH) among young obese Mexican is high. Furthermore, genetic predisposition is a key factor in weight gain and disrupts metabolism. Herein, we used Whole-Exome Sequencing to identify potential causal variants and the biological processes that lead to obesity with progression to NASH among Mexican patients.
Materials and MethodsWhole-Exome Sequencing was performed in nine obese patients with NASH diagnosis with a BMI ≥30 kg/m2 and one control (BMI=24.2 kg/m2) by using the Ion S5TM platform. Genetic variants were determined by Ion Reporter software. Enriched GO biological set genes were identified by the WebGestalt tool. Genetic variants within ≥2 obese NASH patients and having scores of SIFT 0.0-0.05 and Polyphen 0.85-1.0 were categorized as pathogenic.
ResultsA total of 1359 variants with a probable pathogenic effect were determined in obese patients with NASH diagnosis. After several filtering steps, the most frequent pathogenic variants found were rs25640-HSD17B4, rs8105737-OR1I1, rs998544-OR5R1, and rs4916685, rs10037067, and rs2366926 in ADGRV1. Notably, the primary biological processes affected by these pathogenic variants were the sensory perception and detection of chemical stimulus pathways in which the olfactory receptor gene family was the most enriched.
ConclusionsVariants in the olfactory receptor genes were highly enriched in Mexican obese patients that progress to NASH and could be potential targets of association studies.
Obesity is a global health problem recognized as a most important contributor to the development of chronic diseases such as type 2 diabetes, fatty liver disease, hypertension, stroke, dementia, obstructive sleep apnea, and several cancers [1]. Nonalcoholic fatty liver disease (NAFLD) encompasses a spectrum of clinic characteristics ranging from benign steatosis to nonalcoholic steatohepatitis (NASH) and cirrhosis [2]. Hepatic and non-hepatic metabolic disturbances triggered by obesity are key mediators for liver fat accumulation, inflammation, and progression to NASH [3]. In recent years, NASH has emerged as the second most common indication for liver transplants in Western countries, and it is reported that it affects 20-25% of the adult population [3,4]. Mexico ranks as a leader in the prevalence of obesity in adults and children. A high prevalence of NASH and abnormal liver stiffness among young obese Mexicans have been reported [5].
The onset and progression of obesity is a complex process involving several biological processes. Metabolic disorders, adipose tissue dysfunction, mitochondrial dysfunction, and microbiota composition are the more studied processes related to obesity [6, 7]. Nevertheless, food perception has emerged as a new mechanism that increases the risk of obesity. Sensory systems, mainly taste and smell, significantly affect food selection and consumption [8]. Neuroimaging studies have revealed aberrant neural mechanisms in obese subjects [9]. Thus, obesity and related complications can emerge from various processes, and more studies are needed to characterize this intricate network.
Identifying variants with a large biological impact may contribute to understanding the pathways leading to obesity with NASH progression. Genome-Wide Association Studies (GWAS) research has identified some risk markers for obesity and liver damage in genes such as FTO and PNPLA3 among Hispanics and worldwide [10–12]. However, GWAS cannot detect rare variants that predispose to diseases [13]. Herein, we used Whole-Exome Sequencing (WES) to identify low-frequency causative variants and the biological processes that lead to obesity with progression to NASH among Mexican patients.
2Materials and Methods2.1Study PopulationObese patients and control subjects were recruited at the Nutrigenetic Clinic of the Department of Genomic Medicine in Hepatology, Civil Hospital of Guadalajara, "Fray Antonio Alcalde" from 2011 to 2015. A total of nine young extreme obese subjects (body mass index [BMI] average of 43.2 kg/m2) and one control (BMI= 24.2 kg/m2) were included in the study. The BMI measurement, liver damage assessment, and clinical and histological examination details to diagnose NASH were previously published [5]. This study was revised and approved by the Ethical Committees of the Civil Hospital of Guadalajara (ID#HC141/09). Written informed consent was obtained from individuals, and the study protocol was carried out according to the ethical guidelines of the 2013 Declaration of Helsinki.
2.2DNA extraction and WESDNA was isolated from fresh whole blood using a commercial kit (PureLinkTM Genomic DNA Mini Kit, cat. no. K1820-00, Invitrogen, Carlsbad, CA, USA) following the manufacturer's instructions. Whole-exome target regions were amplified using 100 ng of genomic DNA through the Ion AmpliseqTM Exome RDY kit (cat. no. A38262, Thermofisher Scientific, Waltham, MA, USA). Individual samples were ligated to specific Ion XpressTM Barcode Adapters (cat. no. 4471250, Thermofisher Scientific). Unamplified libraries were removed from the samples using the AMPureTM XP reagent (cat. no. A63880, Beckman Coulter, Brea, CA, USA) and freshly prepared 70% ethanol. The library was amplified and purified again by two-round steps with the Agencourt AMPureTM XP reagent. Then, the distribution of the fragments contained in the library was checked on the Agilent 2100 Bioanalyzer instrument (cat. no. G2939A, Agilent Technologies, Santa Clara, CA, USA) using the Agilent High Sensitivity DNA kit (cat. no. 5067-4626, Agilent Technologies). Libraries with a narrow 200 bp length dispersion were quantified through StepOnePlus™ Real-Time PCR System (cat. no. 4376600, Thermofisher Scientific) with the Ion Library TaqMan® Quantitation Kit (cat. no. 4468802, ThermoFisher Scientific) and diluted to 70 pM final concentration. Four barcode exome libraries were combined in equimolar proportions, and template preparations were amplified with the Ion Chef™ System (cat. no. A30011, Thermofisher Scientific) using the Ion 540TM Kit Ion-Chef reagents (cat. no. A30011, Thermofisher Scientific). Enrichment libraries were loaded on Ion 540TM Chips (cat. no. A27766, Thermofisher Scientific), and the sequencing runs were carried out on the Ion S5TM system (cat. no. A27212, Thermofisher Scientific).
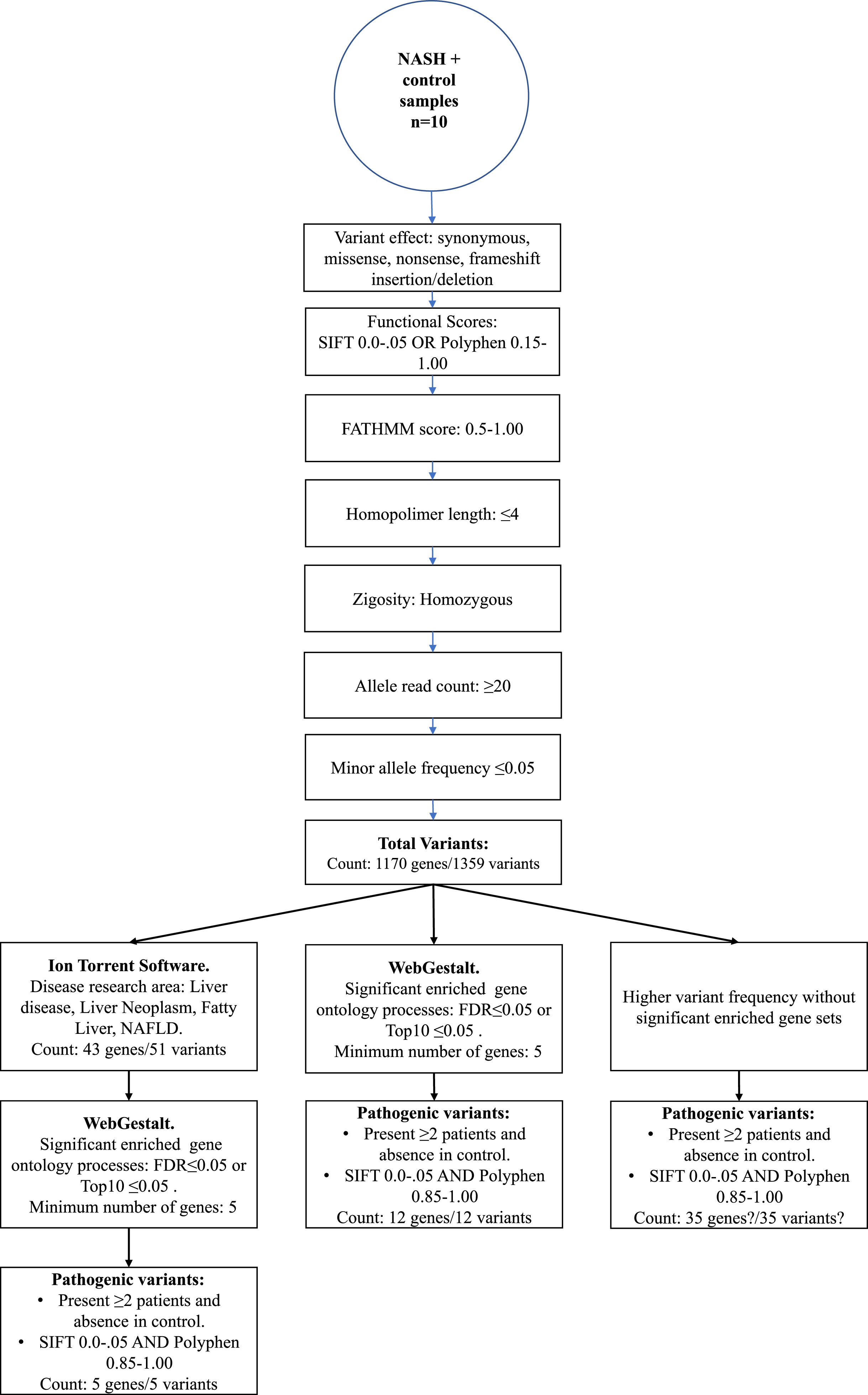
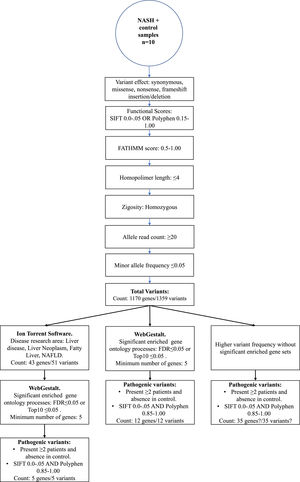
2.3WES data analysisThe Torrent Suite Software V5.12.2 processed the run summary data of each Ion 540 chip. Patients and control sequencing files were aligned to the human reference genome hg19 version, and coverage analysis was targeted to Ampliseq Exome-designed regions. The AmpliSeq Exome files (.BAM files) were uploaded from the Ion Torrent server to the cloud-based Ion ReporterTM software 5.18.20 version. In the last, variant call process in every sample was done with the AmpliSeq Exome single sample (germline) workflow. Then, as depicted in Figure 1, a series of consecutive filter chain parameters were applied in Ion Reporter software to filter the most biological significant variants in each sample. These parameters are typically used in medical investigations to discover new possible variants that confer disease susceptibility. Herein, only variants presented in two or more patients and absence in the control sample were selected to obtain the biological role by functional enrichment analysis web tools.
2.4WEB-based Gene SeT AnaLysis Toolkit (WebGestalt)The list of selected variants/genes filtered in by the Ion Reporter software was introduced in the WebGestalt tool [14] to determine the main gene ontology (GO) processes enriched in obese patients with NASH diagnosis. The most significant enriched gene set ratios with a minimum of 5 genes were determined by the False Discovery Rate (FDR) threshold and Top categories methods with a p-value ≤0.05. The results were shown as tables and volcano plot graphics.
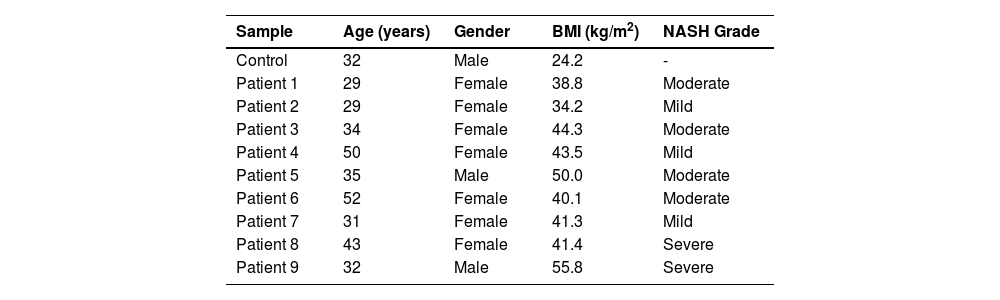
3Results3.1Patients' DataWe performed WES on nine young mestizo Mexican patients with extreme obesity and diagnosis of NASH. A total of 77.8% were female and 22.2% were male, the mean BMI was 43.2 kg/m2 (range 34-56 kg/m2), and patients were mainly young (37.2 ± 8.8 years). The grades of NASH were mainly moderate at 44.4%, mild at 33.3% and severe at 22.2%. The general characteristics of the patients and control are given in Table 1.
Demographic data of the study population
BMI: body mass index; NASH: nonalcohlic steatohepatitis
Average mapped reads in targeted exome regions in the nine patients and the control were 23 million reads, with an average mapped read on the targeted exome of 94%, uniformity of 94%, and mean depth of 72X. The Whole-exome workflow analysis identified ∼53.6 thousand variants in each patient and control sample.
3.3WES identified variantsIon Torrent software annotates the differentially genetic composition between each sample compared to the human genome reference. Thus, different filter chains were necessary to discriminate from neutral/benign to pathogenic variants. In this study, we used three final filters to determine the most probable variants supporting NASH progression. First, through Ion Torrent Software, the standard filter chain was applied to determine variants with probable effects on protein function (blue arrows, Figure 1). A total of 1359 variants in 1170 genes were identified in extreme obese/NASH patients that were not presented in the control sample.
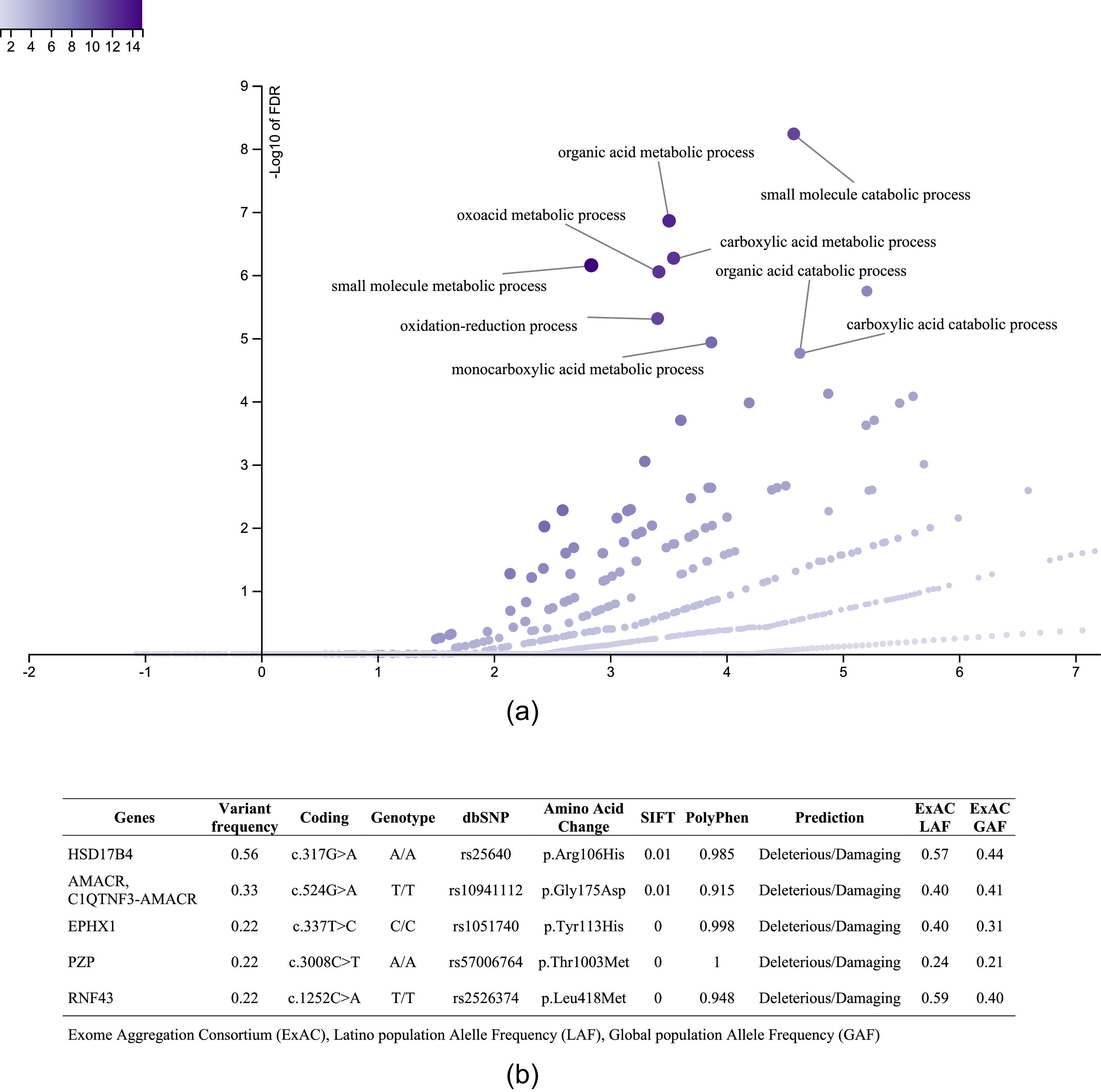
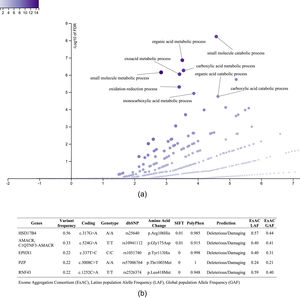
3.3.1Enriched biological processes and variants determined in genes associated with liver diseaseThe 1359 variants were tested in the Disease Research Area (DRA) annotation source database to detect only the variants associated with liver disease. A total of 43 genes and 51 variants were detected. Then, the gene symbols were submitted on the WebGestalt tool to determine the significantly enriched gene ontology biological processes. The top 10 enriched biological processes were highlighted in a volcano plot in Figure 2A. Of these, the leading three gene sets, according to their statistical FDR p-value, were: the small molecule catabolic process (GO:44282), organic acid metabolic process (GO:60802), and carboxylic acid metabolic process (GO:19752). The complete description of the top 10 gene sets enriched as well as the FDR p values are in Supplementary Table 1. We identified five variants that presented SIFT and Polyphen Scores as deleterious/damaging effects on protein function (Figure 2B).
Top 10 enriched biological process and variants determined in genes associated with liver disease. A. Volcano plot that highlights the significantly FDR p value (vertical line) versus the enrichment ratio of a specific gene set (horizontal line). B. List of the variants with deleterious/damaging (based on SIFT and PolyPhen scores) effect in the protein function. The Exome Aggregation Consortium (ExAC), Latino population Alelle Frequency (LAF), Global population Allele Frequency (GAF).
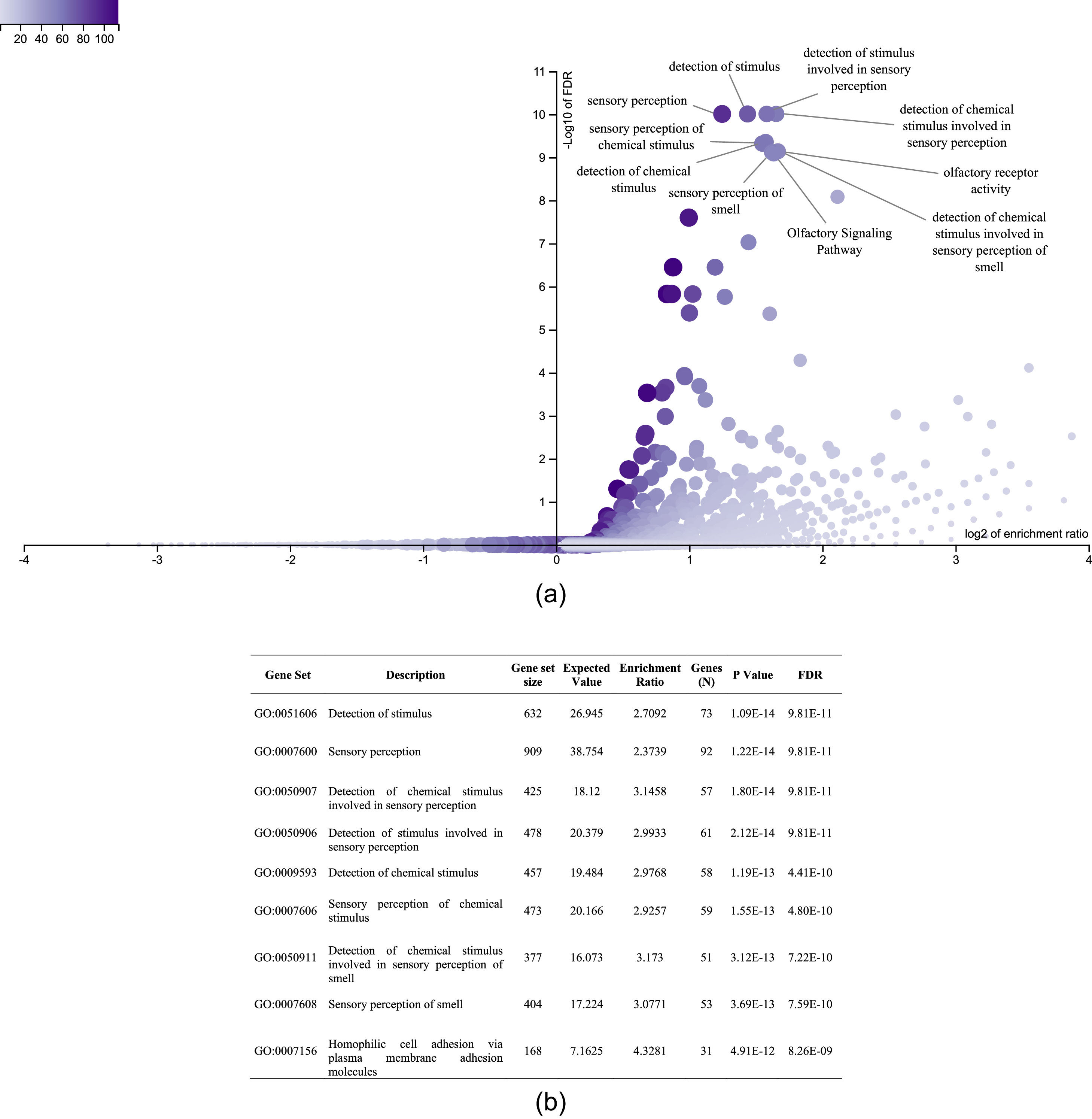
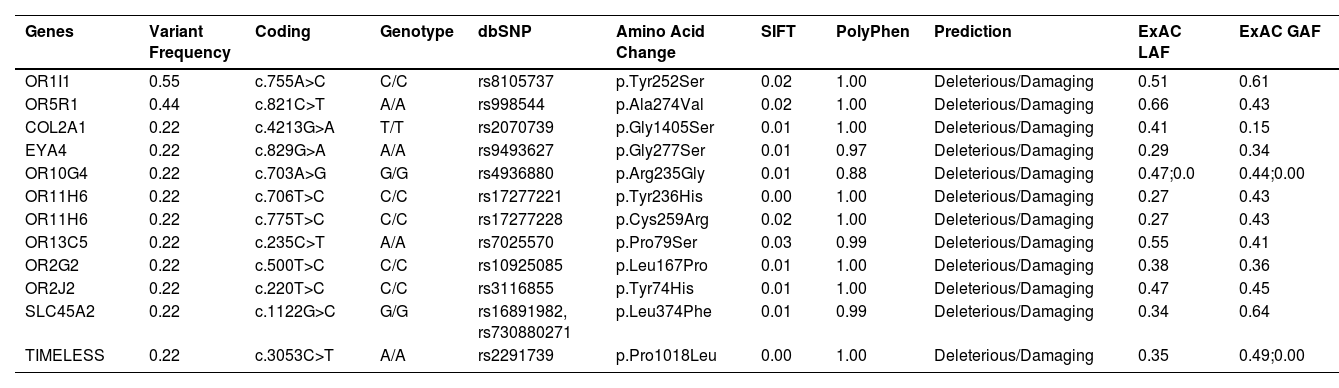
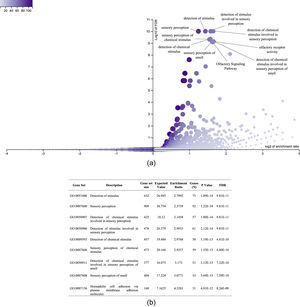
It was also interesting to identify all the biological processes related to the extreme obesity/NASH phenotype, other than those associated with liver disease. We introduced the 1359 variants directly to the WebGestalt tool for this aim. A total of 20 biological processes were statistically significantly enriched by the FDR p<0.05 test. As shown by the volcano plot (Figure 3A), the Top 10 biological processes were detection of stimulus, sensory perception, or nervous system processes. The list of these processes is shown in Figure 3B. After identifying the processes, we looked for the key genes regulating them. Interestingly, the gene family of olfactory receptors (OR) contains 51 members. These genes were associated with the GO molecular function of OR activity GO:0004984 (p=3.1197e-13, FDR=7.2198e-10). Among them, we identified 12 variants that presented SIFT and Polyphen Scores categorized as deleterious/damaging effects on protein function (Table 2). Particularly, rs8105737 (OR1I1) and rs998544 (OR5R1) were present in 56% and 44% of obese/NASH patients.
Top 10 biological process significantly enriched in extreme obese patients with NASH diagnosis. A. Volcano plot highlights the top 10 gene sets, the FDR p value is shown in vertical lines versus the enrichment ratio in the horizontal line.B. List of the top 10 biological processes enriched in obese patients with NASH diagnosis.
Molecular information of pathogenic variants in genes related to olfactory system or nervous system process more frequently presented in extreme obese/NASH patients
Exome Aggregation Consortium (ExAC), Latino population Alelle Frequency (LAF), Global population Allele Frequency (GAF)
Finally, the third filter chain aimed to identify the more frequent variants presented in extreme obese/NASH patients without any biological group association. The list of the complete variants presented in at least two extreme obese/NASH patients and with scores categorized as pathogenic is shown in Supplementary Table 2. A total of 83 new variants were identified that were not previously obtained. For example, gen ADGRV1, a G-protein coupled receptor that binds calcium and is expressed in the central nervous system, had three mutations, rs4916685, rs10037067, and rs2366926. All of them were present in 67% of patients.
4DiscussionObesity is a global health problem that triggers fat liver accumulation. Fatty liver, denoted as simple steatosis (>5% of hepatocytes), can progress to liver inflammation and hepatocytes degeneration leading to the development of NASH [15]. Obesity exhibits a strong heritability component being highly frequent among Hispanic descendent populations [16]. The genes and biological processes involved in obesity and liver complications are diverse. Mexico ranks as a leader in the prevalence of obesity in adults and children [17]. The prevalence of NASH and abnormal liver stiffness among young obese Mexican patients has been reported as high (57%) [5]. Additionally, 3.7% of the general population present extreme obesity [18], associated with liver damage, diabetes, arterial hypertension, and increased mortality [19, 20]; however, the genetic basis of this condition is not entirely known. Herein, we used WES to identify low-frequency causative variants and the biological processes that lead to extreme obesity with progression to NASH among Mexican patients.
After using chain filters to select only genes involved in liver diseases (Figure 1), we could identify the top 10 GO biological processes enriched in patients with obesity and diagnostic of NASH that were previously related to liver disease. As shown in the volcano plot, the main biological processes significantly enriched were metabolic and catabolic. Of the genes involved in these processes, a total of five variants had the most deleterious/damaging predicted effect on the protein function (Figure 2B). 17β-Hydroxysteroid dehydrogenase type 4 (HSD17B4) is a peroxisomal enzyme regulated by phosphatidylserine that controls all androgens and estrogens metabolism [21]. The participation of the HSD17B4 protein in liver cancer proliferation and liver inflammation via STAT3 activation has been described [22]. However, the effect of the variant rs25640HSD17B4 on the expression of the gene is unknown. We found the rs10941112 variant of the α-Methylacyl-CoA racemase (AMACR) gene enriched in patients with obesity and progression to NASH. AMACR catalyzes the degradation of branched-chain fatty acids linking lipid metabolism with nuclear receptors such as FXR and PPAR [23,24]. AMACR is recognized as a cancer marker, but its participation as a liver cancer marker is unclear. Therefore, further investigation should be carried out to identify a possible susceptibility to liver damage and hepatocellular carcinoma in Mexico. Microsomal epoxide hydrolase 1 (EPHX1) regulates the hydrolysis of epoxides and is implicated in adipocyte differentiation [25]. The rs1051740EPHX Tyr113His polymorphism has been described as a possible risk marker for type 2 diabetes and insulin resistance [26]. However, the effect of this variant on the protein function was recently evaluated, and no effect was observed [25]. Thus, further studies are needed to clarify its participation in the establishment of NASH.
On the other hand, pregnancy zone protein (PZP) is a member of the proteinase inhibitor I39 that modulates T-helper cells during proinflammatory processes. Recently, PZP was associated with cell proliferation, invasion, and migration in hepatocellular carcinoma [27]. To the best of our knowledge, the impact of the rs57006764PZP variant on the protein function or its participation in NASH is unknown. Additionally, the Ring finger protein 43 (RNF43) negatively regulates the WNT pathway, thus affecting many cellular processes. A recent study in mice demonstrated that the loss of the RNF43 activity resulted in steatohepatitis and increased unsaturated liver lipids without dietary fat supplementation. Also, RNF43 predisposes liver cancer by controlling proliferation/differentiation and lipid metabolic process in hepatocytes [28]. More studies are needed to characterize the effect of the rs2526374RNF43 variant and its implication in the protein's function. Given that 100% of our patient group had NASH diagnoses, it is expected that the variants could be involved in hepatocyte damage. As the prevalence of risk for NASH is very high among the young Mexican population [5], it might indicate a high susceptibility to developing carcinoma hepatocellular in NASH patients.
The etiology, onset, and progression of NASH are multifactorial. Therefore, we performed a second pipeline to identify all the genes and processes enriched in obese/NASH patients beyond those involved in hepatic diseases. We found sensory perception and detection of chemical stimuli as the main affected processes. Among external senses, olfactory and gustatory systems are the central chemosensory systems in humans that detect environmental odorant and non-odorant cues within food, thus regulating long-term body weight [29,30]. Regarding the gustatory system, a previous study in the Mexican population determined the association of sweet taste receptor 2 Val119Val polymorphism with increased carbohydrate intake and the presence of hypertriglyceridemia [31].
In this study, we identify the OR family as the main contributor to the presence of extreme obesity accompanied by NASH. The olfactory system is related to several functions, including food intake, choice of food, appetite, and satiety mechanisms [32]. This result is in line with a previous WES study in extremely obese Caucasian patients that found a genetic predisposition related to OR genes [33]. Moreover, OR gene methylation marks in obese patients were associated with BMI, waist circumference, and daily intakes of total energy, carbohydrates, protein, and fat [34]. OR genes are expressed in olfactory and non-olfactory tissues such as adipose tissue, liver, kidney, heart, lung, and skin [35–39]. Recent findings demonstrate that some variants in OR genes can regulate hepatic glucose metabolism via the recently discovered adipokine asprosin that regulates glucose production and stimulate appetite [40]. One variant in the OR5R1 gene was associated with metabolic syndrome in the United Kingdom [41]. The rs998544OR5R1 found in this study may have a similar effect, but further studies are needed. We could not find any reported relationship between the widespread variant rs8105737 OR1I1 and liver complications.
On the other hand, in the present study, the rs4916685, rs10037067, and rs2366926 in the Adhesion G protein-coupled receptor V1 (ADGRV1) gene were highly frequent in extremely obese/NASH patients. ADGRV1 protein is expressed in the brain, endocrine tissues, liver, and others, according to the Human Protein Atlas Database (www.proteinatlas.org/ENSG00000164199-ADGRV1/tissue) [42]. Recently, ADGRV1 was detected as one of the most mutated genes in plasma biopsies from patients with hepatocellular carcinoma, and these were presumably critical alterations for protein function [43]. However, the effect of these variants on the progression of NASH is still unknown.
One of the main polymorphisms associated with obesity and liver damage is the Patatin-like phospholipase domain-containing protein 3 (PNPLA3) I148M [44]. Notably, this variant was present in 44% of the study patients. However, it was ruled out for further analysis due to its presence in the control subject. Native Mexicans have the highest PNPLA3 148M risk allele frequency and it is highly prevalent in the mestizo population [45]. These results confirm the high prevalence of this polymorphism in the Mexican population with and without obesity. Thus, the development of obesity and liver damage in Mexico could be conditioned by environmental factors such as food availability, socioeconomics, and other not previously described mutations/polymorphisms with implications within the central nervous system or food perception as the OR genes found in this study.
The results of the present study point out the main biological processes affected in extreme obesity with progression to NASH and variants present in coding sequences of the genome that have the most pathogenic predicted effect on the protein function. However, this study presents some limitations. First, lower frequency variants could have gone unnoticed due to the low number of samples available for the study. Second, the nine extremely obese patients were referred to the Nutrigenetic Clinic after bariatric surgery, so biochemical parameters were unavailable to discuss. Third, further studies with a larger sample size are needed to validate the variants' effect on metabolic and liver function tests.
5ConclusionsObesity and its liver complications represent essential health and economic burden in many countries. With the alarming increase in fat and sugar consumption worldwide, identifying those individuals with genetic susceptibility to develop obesity/extreme obesity with progression to liver damage might facilitate the implementation of prevention strategies in the population at risk. Variants in OR genes were highly enriched in Mexican extreme obese patients that progress to NASH and could be potential targets of association studies. Variants in genes related to carcinogenic processes give additional risk to extreme obese/NASH patients to develop liver cancer. This study paves the way for understanding the genes and biological processes leading to extreme obesity and NASH in the Mexican population and provides an approach for implementing precision medicine.
FundingThis work was supported by Programa para el Desarrollo Profesional Docente (PRODEP) to Torres-Reyes LA, No. UDG-PTC-1490; Programa para el Desarrollo Profesional Docente (PRODEP) to Gonzalez-Aldaco K, No. UDG-PTC-1422; Programa de Fortalecimiento de Investigación y Posgrado -Universidad de Guadalajara and partially by Grant PN-2017-5254-CONACYT to Panduro A. Fundación Clínica Médica Sur provided the resources for the publication of this research.
Author contributionsRoman S and Torres-Reyes LA conceived the study; Panduro A clinically evaluated all patients and validated clinical data. Torres-Reyes LA and Gonzalez-Aldaco K carried out experimental work and drafted the manuscript. Torres-Reyes LA did formal analysis, data curation, resources and bioinformatic analysis. Jose-Abrego A validated bioinformatic data and analysis. Roman S revised all experimental data and copyedited the English version. All authors contributed intelligently, critically revised, and approved the final version of the manuscript. Roman S is responsible for the integrity of the work as a whole.