Long non-coding RNA (lncRNA) EPIC1 (epigenetically-induced lncRNA1) is likely involved in human cancer by promoting cell cycle progression. Our study was carried out to investigate the involvement of EPIC1 in gallbladder cancer (GBC).
MethodsExpression levels of EPIC1 in two types of tissues (GBC and paracancerous) and plasma were measured by performing qPCR. GBC-SD and SGC-996 cells were transfected with low expression in tumor (LET) and EPIC1 expression vectors.
ResultsThe present study found that EPIC1 was upregulated in tumor tissues than in paracancerous tissues of GBC patients, and plasma levels of EPIC1 were significantly correlated with levels of EPIC1 in tumor tissues. LncRNA LET was downregulated in tumor tissues than in paracancerous tissues and was inversely correlated with EPIC1 in both tumor tissues and paracancerous tissues. Overexpression of EPIC1 led to downregulated LET, and LET overexpression also mediated the downregulation of EPIC1. EPIC1 led to accelerated GBC cell proliferation and inhibited apoptosis. Overexpression of LET played opposites roles. In addition, LET overexpression attenuated the effects of EPIC1 overexpression on cancer cell proliferation and apoptosis.
ConclusionsLncRNA EPIC1 promoted proliferation and inhibited apoptosis of GBC cells by interacting with LET.
As a common malignancy originated in the biliary tract, gallbladder cancer (GBC) mainly affects females and elders [1, 2]. The incidence of GBC is not high in the United States, only affecting about 1000 Americans and causing about 500 deaths every year [3]. However, in China, GBC occurs in 3-5 out of 100,000 people, with a mortality >70% [4]. The occurrence of GBC is closely related to the existence of gallstones [5], while only 1/200 of gallstone patients will develop GBC [1]. At present, therapeutic methods for GBC mainly included surgical resection, adjuvant chemotherapy, and immunotherapy [6–8]. However, the pathogenesis of GBC remains poorly understood [5,9], which is a challenge for the development of novel therapeutic approaches.
It has been well-established that long (>200nt) non-coding RNAs (lncRNAs) contribute significantly to the development and progression of GBC [9,10]. Different from mRNAs as the template for protein synthesis, lncRNAs participate in cancer biology mainly by regulating the expression of certain oncogenes and tumor suppressors [10–12]. A better understanding of the roles of lncRNAs may provide new insights into cancer prevention and treatment [13]. However, the functionality of most lncRNAs remains unknown. The epigenetically-induced lncRNA1 (EPIC1) has been characterized as an oncogenic lncRNA in many types of cancers [11,14,15], whereas its involvement in GBC is unclear. Our preliminary deep sequencing data showed that EPIC1 was downregulated in GBC and inversely correlated with the lncRNA low expression in tumor (LET), which has been characterized as a tumor suppressor in GBC [16]. This study aimed to investigate the potential interaction between EPIC1 and LET in GBC.
2Methods2.1SubjectsA total of 60 GBC patients (26 males and 34 females; age of 58 to 79 years, 69.3±6.6 years) were enrolled in the study. These patients were selected from the 133 GBC patients admitted to Hubei Provincial Cancer Hospital between May 2015 and January 2018 if they were newly diagnosed GBC and had not been retreated previously. Patients were excluded if they were 1) recurrent cases, 2) transferred from other hospitals, and 3) complicated with other severe clinical disorders. Among the enrolled patients, 12, 13, 18, and 17 cases were at stages I-IV, respectively, based on their clinical findings according to AJCC staging methods. This study was approved by the Ethics Committee of Hubei Provincial Cancer Hospital. All patients were informed of the experimental details and signed the informed consent.
2.2GBC specimens and cellsBefore any therapies, paracancerous and GBC tissues were collected from all GBC patients through biopsy. All tissues were subjected to pathological examinations and confirmed to be correctly diagnosed.
Fasting blood (3ml) was extracted from all GBC patients before any therapies and added into the EDTA-treated tubes. After centrifugation at 1200 g for 15min, supernatant (plasma) was collected.
Human GBC cell lines GBC-SD and SGC-996 from the Cell Bank of Chinese Academy of Sciences (Shanghai, China) were cultured in DMEM medium supplemented with 10% fetal bovine serum (FBS) at 37°C in a humidified incubator with 5% CO2.
2.3Vectors and transient transfectionsEPIC1 and LET expression vectors were constructed using pcDNA3 as the backbone (RIBOBIO, Guangzhou, China). GBC-SD and SGC-996 were harvested, counted, and transiently transfected with 10 nM expression or empty pcDNA3 vectors per 106 cells using Lipofectamine 2000 (Invitrogen, USA). Control (C) cells were cells without transfections. At 24 h of post-transfection, cells were collected and used for future experiments.
2.4Total RNA extractionTissues were ground in liquid nitrogen. Total RNAs in 0.01 g tissues, 0.2 ml plasma, and 106 cells were extracted using Trizol reagent (Invitrogen, USA). RNA samples were precipitated and washed using 75% Ethanol. All RNA samples were stored in liquid nitrogen before any experiment.
2.5QPCRTotal RNA samples were digested with DNase I for 1 h at 37°C to remove genomic DNAs. The digested RNA samples were used as templates in reverse transcription using the AMV reverse transcriptase (GIBCO, USA). The levels of EPIC1 and LET were measured using QuantiTect SYBR Green PCR Kit (Qiagen) with GAPDH as the internal control and quantified using the 2−ΔΔCt method. All experiments were performed in triplicates.
2.6Cell proliferation analysisCells transfected with EPIC1 or LET were counted, prepared as single cell suspension with 3 × 104 cells per ml DMEM medium containing 10% FBS, loaded into 96-well cell culture plate with 0.1 ml per well, and cultured at 37°C in a humidified incubator with 5% CO2. Three replicate wells were set for each transfection group. Following that, 10 μl CCK-8 solution (Sigma-Aldrich) was added into each well at 2h before the termination of cell culture. After cell culture was ended, 10 μl DMSO was added into each well, and the OD values were measured at 450 nm.
2.7Cell apoptosis analysisCells transfected with EPIC1 or LET were counted, prepared as single cell suspension with 3 × 104 cells per ml DMEM medium containing 10% FBS, loaded into 6-well plates with 2 ml per well, and cultured at 37°C in a humidified incubator with 5% CO2 for 48 h. Three replicate wells were set for each transfection group. After that, cells were digested with 0.25% trypsin, stained with propidium iodide (PI) and Annexin V-FITC (Dojindo, Japan), and subjected to flow cytometric analyses to separate apoptotic cells.
2.8Statistical analysisAll qPCR, cell proliferation, and cell apoptosis experiments were repeated 3 times. Data were expressed as the mean values. Differences were explored by either paired t test (GBC vs. paracancerous tissues) or one-way ANOVA combined with Tukey test (among different cell transfection groups). Correlations were analyzed by linear regression. A p<0.05 was considered statistically significant.
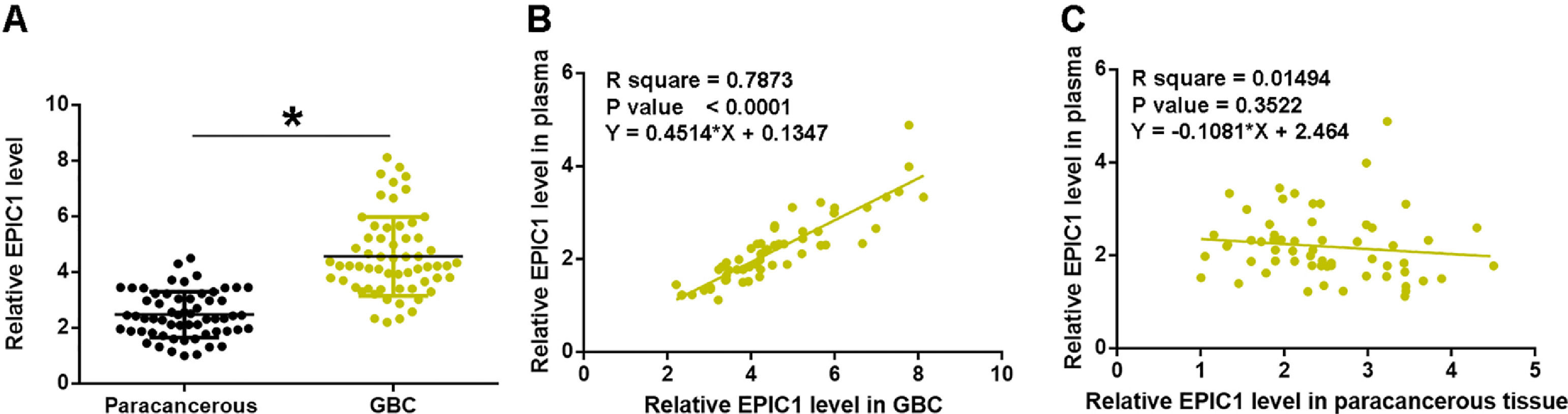
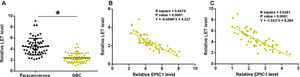
3Results3.1EPIC1 was upregulated in GBC and positively correlated with its level in plasmaEPIC1 levels in GBC and paracancerous tissues and plasma were measured by performing qPCR. The differences in EPIC1 levels between GBC and paracancerous tissues were explored by paired t test. Compared to paracancerous tissues, EPIC1 level was significantly higher in the GBC tissues (Fig. 1A, p<0.05). Correlations between EPIC1 levels in GBC and paracancerous tissues and in plasma were analyzed by the linear regression. It was observed that plasma EPIC1 level was significantly correlated with EPIC1 level in the GBC tissues (Fig. 1B) but not in the paracancerous tissues (Fig. 1C).
EPIC1 was upregulated in GBC and positively correlated with its level in plasma.
EPIC1 levels in GBC and paracancerous tissues and in plasma were measured by qPCR. Difference in EPIC1expression levels between GBC and paracancerous tissues was analyzed in paired t test (A). Correlations between EPIC1 levels in GBC (A) and paracancerous (B) tissues and in plasma were analyzed by linear regression. qPCR was performed 3 times and data are presented as the mean values. * p<0.05.
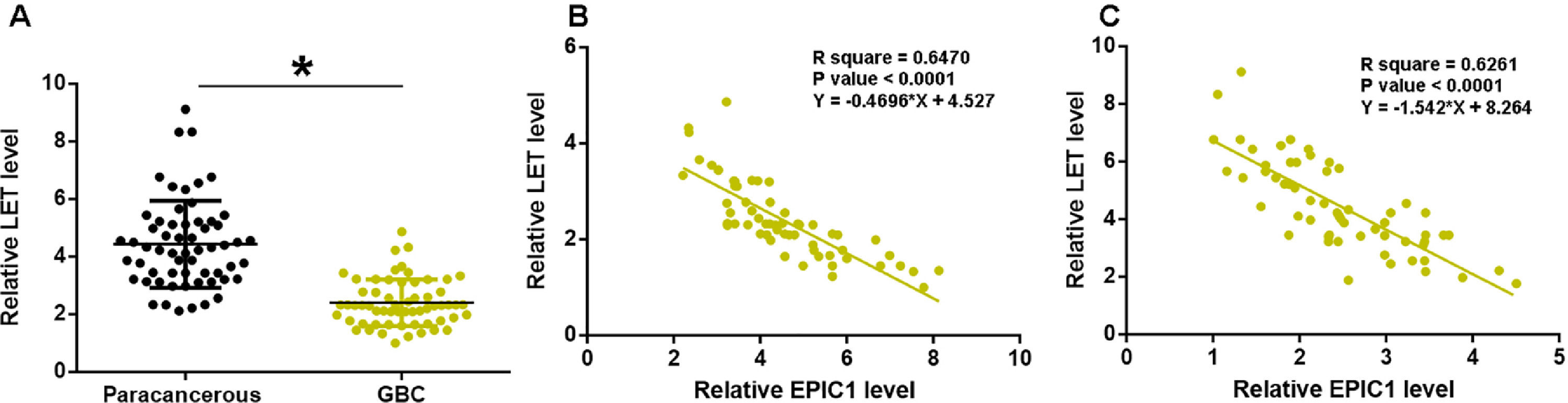
LET levels in GBC and paracancerous tissues were measured by qPCR. The difference in LET levels between the GBC and paracancerous tissues was explored by paired t test. Compared to paracancerous tissues, LET expression level was significantly lower in GBC tissues (Fig. 2A, p<0.05). Correlation between EPIC1 and LET was analyzed by linear regression. It was observed that LET was inversely correlated with EPIC1 in both GBC (Fig. 2B) and paracancerous tissues (Fig. 2C).
LET level was downregulated in GBC and was inversely correlated with EPIC1 level.
LET expression in GBC and paracancerous tissues was measured by qPCR. Differences in LET expression levels between GBC and paracancerous tissues were explored by paired t test (A). Correlations between EPIC1 and LET levels in both tumor tissues (B) and paracancerous tissues (C) were analyzed by linear regression. qPCR was performed 3 times and data are presented as the mean values. * p<0.05.
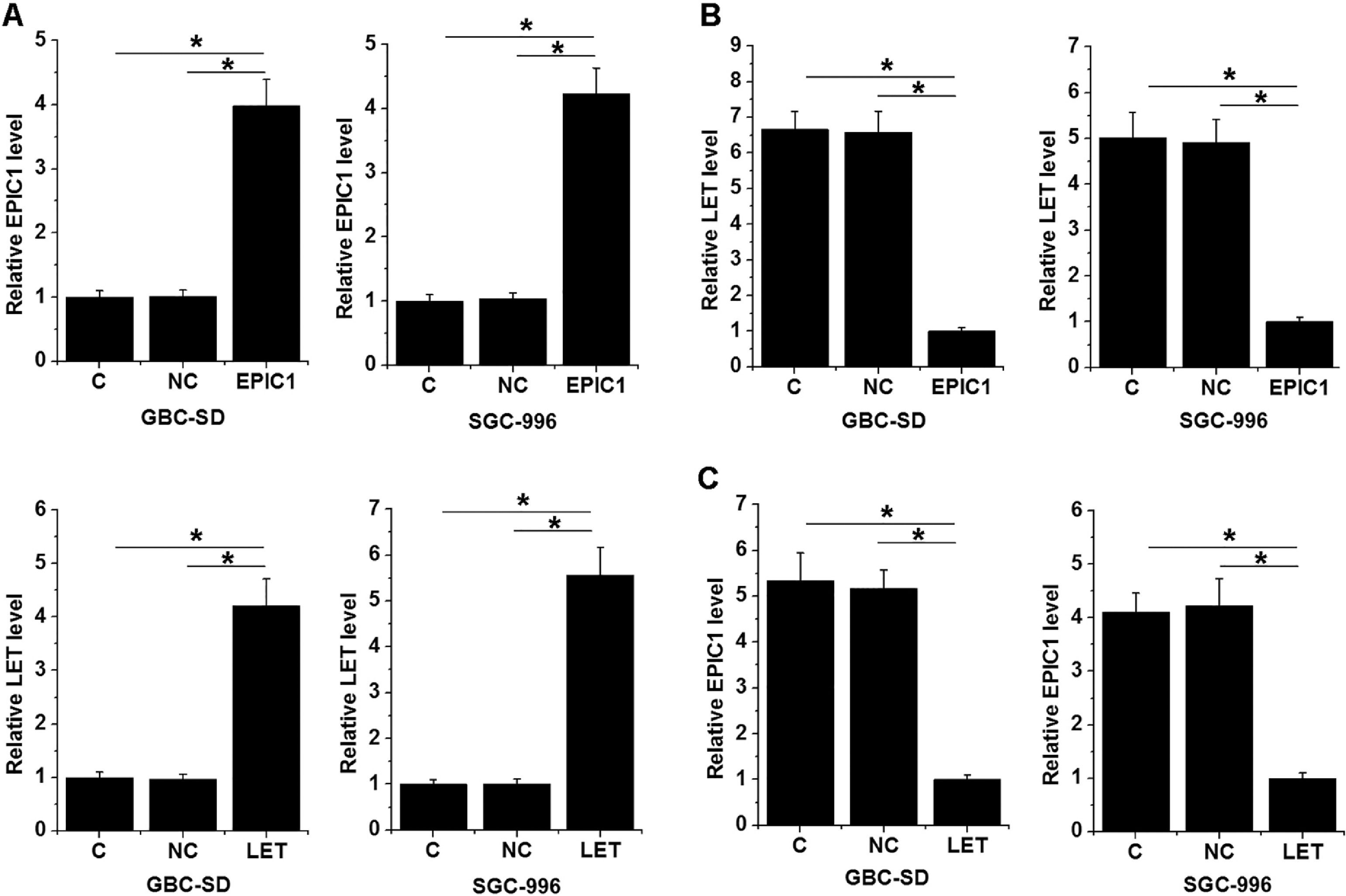
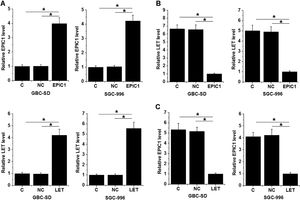
GBC-SD and SGC-996 cells were transfected with LET and EPIC1 expression vectors. At 24 h of post-transfections, LET and EPIC1 levels were significantly increased in LET and EPIC1 overexpressed cells, respectively, compared with the C and NC (empty vector) cells (Fig. 3A, p<0.05). In addition, EPIC1overexpression led to downregulated LET level (Fig. 3B) and LET overexpression also downregulated EPIC1 level (Fig. 3C) in both cell lines (p<0.05).
LET and EPIC1 negatively regulated each other in GBC tissues.
GBC-SD and SGC-996 cells were transfected with LET and EPIC1 expression vectors. At 24h of post-transfections, LET and EPIC1 overexpression were confirmed by qPCR (Fig. 3A, p<0.05). The effects of EPIC1 overexpression on LET (B) and the effects of LET overexpression on EPIC1 (Fig. 3C) were explored by qPCR. All experiments were repeated 3 times and all data are presented as the mean values. * p<0.05.
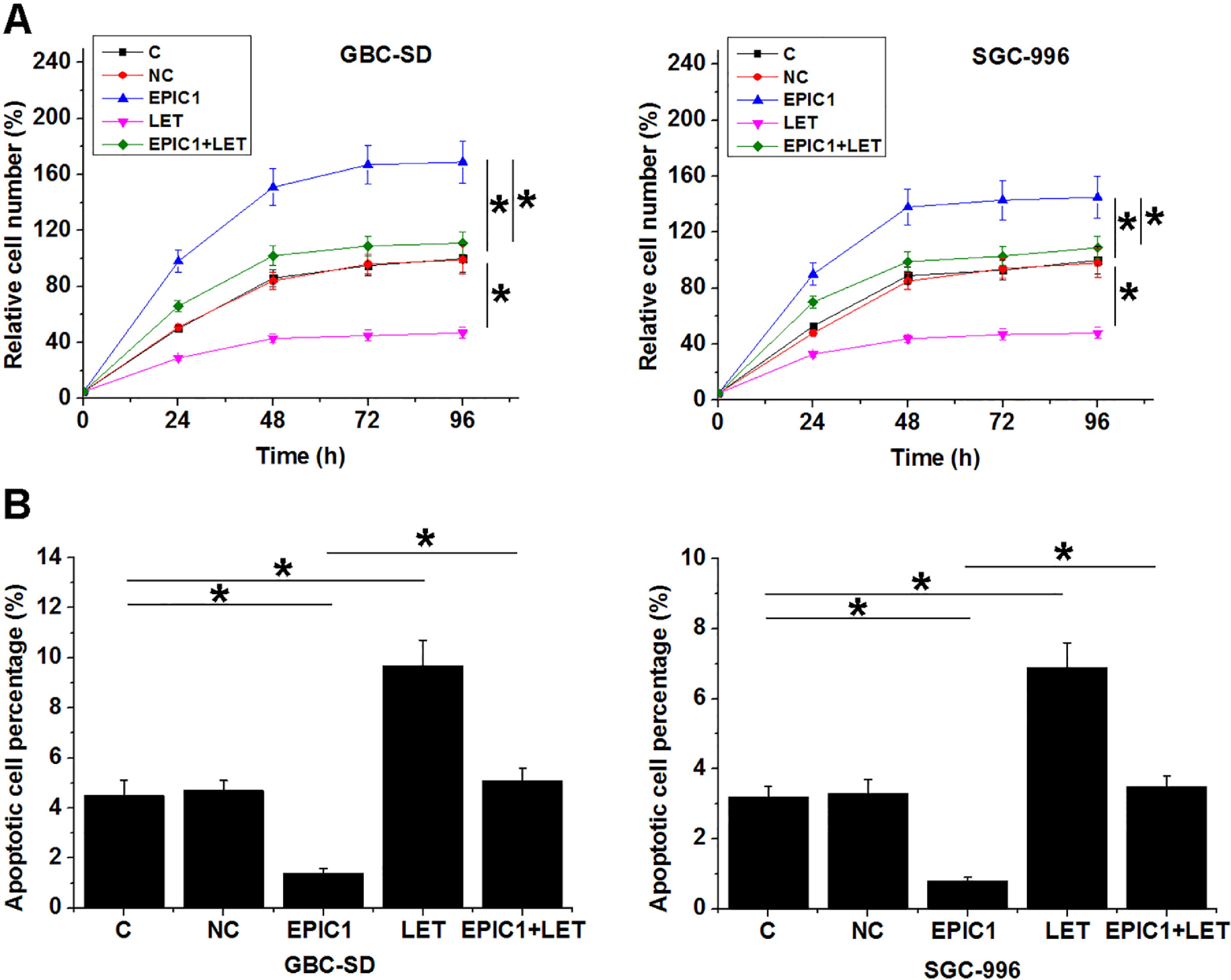
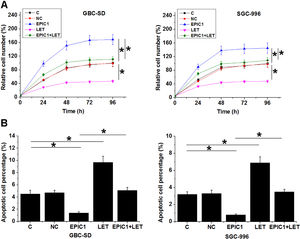
Compared to the C and NC groups, EPIC1 overexpression increased proliferation rate (Fig. 4A) and reduced apoptotic rate (Fig. 4B) of both GBC-SD and SGC-996 cells (p<0.05), while LET overexpression played opposites roles. Moreover, LET overexpression also attenuated the effects of EPIC1 overexpression on cancer cell proliferation and apoptosis.
The interaction between EPIC1 and LET in regulating GBC cell proliferation and apoptosis.
The effects of EPIC1 and LET overexpression on the proliferation and apoptosis of both GBC-SD and SGC-996 cells were analyzed by cell proliferation (A) and apoptosis (B) assays. All experiments were repeated 3 times and all data are presented as the values. *p<0.05.
This study analyzed the interactions between lncRNAs EPIC1 and LET in GBC. The results showed that EPIC1 was upregulated and LET was downregulated in GBC. In addition, EPIC1 and LET formed a negative feedback regulation loop to regulate the apoptosis and proliferation of GBC cells.
The function of EPIC1 has been studied in many different types of cancers. Most studies supported the role of EPIC1 as an oncogenic lncRNA in cancer biology [12,15,16]. For instance, EPIC1 promoted cell cycle progression via interacting with MYC [15]. EPIC1 was upregulated in lung cancer, and EPIC1 overexpression enhanced cell proliferation [14]. However, Zhao et al. recently reported that EPICA promoted MEF2D ubiquitylation to suppress cancer cell invasion and viability [17], indicating it functions as a tumor suppressor in this disease. Therefore, EPIC1 may have different functions in different types of cancers. Our study is the first to report EPIC1 overexpression in GBC. We also showed that EPIC1 overexpression enhanced proliferation and inhibited apoptosis of GBC cells. Our data suggest EPIC1 as an oncogenic lncRNA in GBC.
LET plays a tumor suppressive role in different types of cancers by regulating multiple cell behaviors, such as proliferation, apoptosis, and invasion [18,19], and LET downregulation predicted poor survival of GBC patients [16]. Our study showed that LET inhibited GBC progression by regulating GBC cell proliferation and apoptosis, further confirming the role of LET as a tumor suppressor in GBC. It is known that LET expression in cancer can be regulated by other lncRNAs, such as lncRNA DANCR. We found that EPIC1 and LET formed a negative feedback regulation loop to regulate GBC cell behaviors. However, the study had several limitations. First, the mechanism underlying the interaction between EPIC1 and LET remains unclear and needs to be further analyzed. Moreover, the current study mainly focused on the effect of EPIC1 expression on the proliferation of GBC cells in vitro. Further studies are required to determine whether similar findings occur in vivo.
In summary, our study is the first report to reveal that EPIC1 is upregulated in GBC tissues and affects GBC progression by regulating LET. Our results provide a theoretical basis for potential GBC treatment.
Ethical approvalThis study was approved by the Ethic Committee of Hubei Provincial Cancer Hospital.
Data availability statementNot applicable.
Author contributionAll authors contributed to data analysis, manuscript drafting and revising, approved the final version to be published, and agreed to be accountable for all aspects of the work.