Editado por: Dra. Núria Torner CIBER Epidemiologia y Salud Publica CIBERESP Unitat de Medicina Preventiva i Salut Pública Departament de Medicina, Universitat de Barcelona
Más datosVaccination is one of the most pertinent prevention strategies for the Coronavirus Disease 2019 (COVID-19) pandemic. Several factors, both intrinsic (particularly genetic) and extrinsic, can influence vaccine efficacy. However, very little research has been conducted into the genetic component's impact on immunogenicity following COVID-19 vaccination. Therefore, we present the antibody formation in thirteen people who received a third vaccination (booster) dose of the Moderna mRNA-1273 vaccine and the differences in the polymorphism Tumor Necrosis Factor-Alpha (TNF-α) related genes in this population.
MethodsOur study included 13 participants with no comorbidities or a history of COVID-19 infection. The Chemiluminescent Microparticle Immunoassay (CMIA) was used to measure antibody production in serum. Polymorphism was recognized using the polymerase chain reaction (PCR) amplification technique.
ResultsIn this study, TNF-α related gene (GG) significantly influenced the formation of the antiSARS-CoV-2 spike protein IgG antibody level (p = 0.005) in our sample.
ConclusionAlthough the polymorphism of the cytokine gene, particularly TNF-α, seems to influence antibody levels in our study population, a more comprehensive analysis is required for better generalization due to the nature of our pilot study.
La vacunación es una de las estrategias de prevención más pertinentes ante la pandemia de Enfermedad del coronavirus 2019 (COVID-19). Varios factores, tanto intrínsecos (particularmente genéticos) como extrínsecos, pueden influir en la eficacia de la vacuna. Sin embargo, se ha realizado muy poca investigación sobre el impacto del componente genético en la inmunogenicidad después de la vacunación contra el COVID-19. Por lo tanto, presentamos la formación de anticuerpos en trece personas que recibieron una tercera dosis de vacunación (refuerzo) de la vacuna Moderna mRNA-1273 y las diferencias en los genes relacionados con el polimorfismo el factor de necrosis tumoral alfa (TNF-α) en esta población.
MétodosNuestro estudio incluyó a 13 participantes sin comorbilidades o antecedentes de infección por COVID-19. Se utilizó el inmunoanálisis de micropartículas quimioluminiscentes (CMIA) para medir la producción de anticuerpos en suero. El polimorfismo se reconoció utilizando la técnica de amplificación de la reacción en cadena de la polimerasa (PCR).
ResultadosEn este estudio, el gen relacionado con TNF-α (GG) influyó significativamente en la formación del nivel de anticuerpos IgG de proteína de punta antiSARS-CoV-2 (p = 0,005) en nuestra muestra.
ConclusionesAunque el polimorfismo del gen de la citoquina, particularmente el TNF-α, parece influir en los niveles de anticuerpos en nuestra población de estudio, se requiere un análisis más completo para una mejor generalización debido a la naturaleza de nuestro estudio piloto.
Vaccines are essential for infection control measures, especially during the current Coronavirus Disease 2019 (COVID-19) pandemic.1 The impact of effective vaccination can be seen either directly or indirectly in reducing new and severe COVID-19 cases, especially following booster vaccination doses.2,3
Individual responses to vaccination can be influenced by intrinsic and extrinsic factors. Numerous intrinsic factors can contribute to it, including age, gender, genetics, and underlying diseases. Meanwhile, some extrinsic factors have been identified, such as environmental factors, lifestyle factors, nutritional factors, and vaccine administrations.4 Polymorphism in several genes, such as Interferon-lambda (IFNL)3/4, interleukin (IL)-6 (influenza), and CD46 (measles) was also found to affect antibody formation.5
As previously stated, genetic factors may be responsible for influencing COVID-19 immunity. Several genes, such as the Human leukocyte antigens (HLA) and Angiotensin-converting enzyme 2 (ACE2) have been found to affect COVID-19 severity and outcome.6 Furthermore, people with the A allele of the Tumor Necrosis Factor-Alpha (TNF-α) polymorphism are more susceptible to COVID-19 infection and severe disease condition.7
The analysis of the genetic component's impact on antibody production after COVID-19 vaccination is sparse. This study aims to identify the genotypic variation of TNF-α genes in the Indonesian population and analyze the antibody titer levels following COVID-19 vaccination.
Materials and methodsStudy overviewThis cohort study was conducted using a longitudinal repeated-measure approach from March 2021 till August 2021. Participants in the study were people aged ≥18 years and recruited based on consecutive sampling method. Several exclusion criteria were applied, including: (1) autoimmune disease, (2) fever at the time of blood sampling, (3) prior SARS-CoV-2 infection, and (4) reactive antigen screening. This study has already got the ethical approval from our institution's ethical review board (approval number: 062–2021).
Study processThe vaccination procedure (conducted in a tertiary-level hospital) was as follows: the first dose was administered in early March 2021, the second dose was administered 14 days later, and the third was administered 20 weeks after the previous dose. After acquiring participants' consent, data on demographic characteristics, comorbidity history, and adverse events following immunization were compiled. Participants' anthropometric data, temperature, blood pressure (BP), and venous blood glucose level were measured.
Antibody detectionVenous blood was drawn thrice (14 days after the first vaccination, 30 days following the second vaccination (all with CoronaVac), and 14 days after the third vaccination (booster) with the Moderna mRNA-1273 vaccine. Antibody level (IgG anti-spike (S)) was measured by using the SARS-CoV-2 IgG II Quant kit (Abbott Diagnostics). The examination was carried out using the Chemiluminescent Microparticle Immunoassay (CMIA) method using 75 μL of serum as the sample. Antibody level was expressed as arbitrary units (AU)/mL.
Polymorphism detectionApart from the antibody measurements, other venous blood samples (whole blood, 200 μL) were drawn for Deoxyribonucleic acid (DNA) isolation. The QIAamp DNA mini kit was then used to process it (Becton Dickinson, USA). The polymerase chain reaction (PCR) amplification technique was used to identify the point polymorphisms.8Table 1 described the primer pair and restriction enzyme used in our specimen.
Primer pair and restriction enzyme utilized in this study.
| Polymorphisms type | Primer pair | Restriction enzyme |
|---|---|---|
| rs12979860 C/T | Forward: 5′-GCTTAT CGCATACGGCTAGG-3′Reverse: 5′-GGCTCAGGGTCAATCACAG-3′ | Bsh1236I (BstUI) |
| rs368234815 TT/DG | Forward: 5′- GACGCAGGACCCCTTGGGACAGGA-3’Reverse: 5′- CTGGGCCGCAGTGGCCGCGAGG-3’ | MspA1 |
| TNF-α-308 G/A | Forward: 5’AGG CAA TAG GTT TTG AGG GCC AT 3’Reverse: 5’TCC TCC CTG CTC CGA TTC 3’ | Nco1 |
The PCR procedure was initiated with a 5-min denaturation step at 95 °C. Sequentially, we rendered denaturation at 95 °C for 45 s (35 cycles), annealing at 60°C for 30 s, extension at 72 °C for 45 s, and final extension at 72 °C for one minute. The amplification results were identified via electrophoresis in 2% agarose gel (30 min). Meanwhile, the Restriction fragment length polymorphism (RFLP) was implemented by incubating the PCR results of each polymorphism overnight with the endonuclease enzyme. After incubation, the restriction results were analyzed using the electrophoresis technique with a 3% agarose gel. The last step of polymorphism identification (by PCR and RFLP method) involved the use of a Gel Doc 1000 machine (Uvitec, UK) to visualize the sample with ultraviolet (UV) light.
Statistical analysisThe data were analyzed using IBM Statistical Package for the Social Sciences (SPSS) Statistics for Windows, version 26.0 (IBM Corp., Armonk, NY, USA). Quantitative variables (such as antibody levels) were presented as mean with standard deviation (SD) or median with interquartile range (IQR) based on data distribution assessed by Shapiro-Wilk test. Additionally, the upsurge in antibody levels between the second and third vaccination dose was assessed using the Paired T-test (or Wilcoxon test for non-parametric assessment).
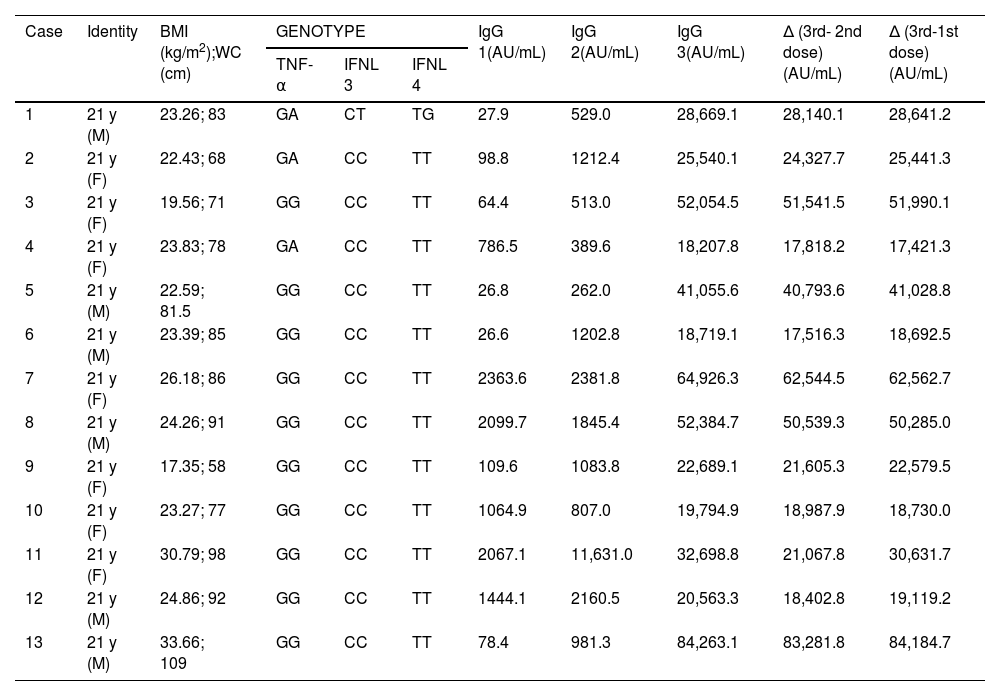
ResultsFifteen participants were enrolled at the start of the study. However, two people contracted COVID-19 after the second vaccination dose and were excluded later. Thirteen cases (7 women and 6 men) were finally selected for the antibody quantification. All participants were at the age of 21. They did not report any known comorbidities. There were no serious side effects observed due to the vaccination process. The blood pressure and blood glucose tests revealed no abnormalities (data not shown). Three participants, however, are obese (body mass index greater than 25 kg/m2) (Table 2).
Participant characteristics.
| Case | Identity | BMI (kg/m2);WC (cm) | GENOTYPE | IgG 1(AU/mL) | IgG 2(AU/mL) | IgG 3(AU/mL) | Δ (3rd- 2nd dose) (AU/mL) | Δ (3rd-1st dose) (AU/mL) | ||
|---|---|---|---|---|---|---|---|---|---|---|
| TNF-α | IFNL 3 | IFNL 4 | ||||||||
| 1 | 21 y (M) | 23.26; 83 | GA | CT | TG | 27.9 | 529.0 | 28,669.1 | 28,140.1 | 28,641.2 |
| 2 | 21 y (F) | 22.43; 68 | GA | CC | TT | 98.8 | 1212.4 | 25,540.1 | 24,327.7 | 25,441.3 |
| 3 | 21 y (F) | 19.56; 71 | GG | CC | TT | 64.4 | 513.0 | 52,054.5 | 51,541.5 | 51,990.1 |
| 4 | 21 y (F) | 23.83; 78 | GA | CC | TT | 786.5 | 389.6 | 18,207.8 | 17,818.2 | 17,421.3 |
| 5 | 21 y (M) | 22.59; 81.5 | GG | CC | TT | 26.8 | 262.0 | 41,055.6 | 40,793.6 | 41,028.8 |
| 6 | 21 y (M) | 23.39; 85 | GG | CC | TT | 26.6 | 1202.8 | 18,719.1 | 17,516.3 | 18,692.5 |
| 7 | 21 y (F) | 26.18; 86 | GG | CC | TT | 2363.6 | 2381.8 | 64,926.3 | 62,544.5 | 62,562.7 |
| 8 | 21 y (M) | 24.26; 91 | GG | CC | TT | 2099.7 | 1845.4 | 52,384.7 | 50,539.3 | 50,285.0 |
| 9 | 21 y (F) | 17.35; 58 | GG | CC | TT | 109.6 | 1083.8 | 22,689.1 | 21,605.3 | 22,579.5 |
| 10 | 21 y (F) | 23.27; 77 | GG | CC | TT | 1064.9 | 807.0 | 19,794.9 | 18,987.9 | 18,730.0 |
| 11 | 21 y (F) | 30.79; 98 | GG | CC | TT | 2067.1 | 11,631.0 | 32,698.8 | 21,067.8 | 30,631.7 |
| 12 | 21 y (M) | 24.86; 92 | GG | CC | TT | 1444.1 | 2160.5 | 20,563.3 | 18,402.8 | 19,119.2 |
| 13 | 21 y (M) | 33.66; 109 | GG | CC | TT | 78.4 | 981.3 | 84,263.1 | 83,281.8 | 84,184.7 |
Abbreviation: BMI, body mass index; IFNL, interferon lambda (λ); TNF, tumor necrosis factor; WC, waist circumference.
Note: IgG 1 = measured 14 days after the first vaccination, IgG 2 = measured 30 days following the second vaccination, IgG 3 = measured14 days after the third vaccination (booster).
We discovered that the polymorphism might influence antibody formation following vaccination, particularly after the third dose (booster) vaccination. All participants had a value greater than the protective humoral response threshold (840 AU/mL) after booster vaccination.
The most significant differences can be found in TNF-α-related polymorphism. After the third vaccination, the anti-S antibody level increased significantly from 1143.30 AU/mL (Interquartile range/IQR, 733.5–2215.8) to 36,877.2 AU/mL (IQR, 20371.2–55,520.1) (p = 0.005) in the sample with GG point polymorphism. Meanwhile, after booster vaccination, the median antibody level for participants with the GA gene increased from 529.0 AU/mL to 25,540 AU/mL (p = 0.109). The GG genotype was found in 76.9% of participants, while the GA genotype was found in 23.1%.
After booster vaccination, the median antibody levels for participants with polymorphism in the IFNL gene were the same for both dominant polymorphism types (CC for IFNL3 gene and TT for IFNL4 gene; 29,119.45 AU/mL) and non-dominant type (CT for IFNL3 gene and TG for IFNL4 gene; 28,669.10 AU/mL). However, because there was only one sample in each CT and TG group, the differences could not be ascertained further.
DiscussionBased on observed polymorphisms of the TNF-α related genes, this study discovered differences in antibody formation after the third dose of the Moderna mRNA-1273 vaccine. Previous research has shown that the Moderna vaccine (booster dose) is essential in eliciting higher antibody levels (by 11.5 to 32 times), regardless of the type of vaccine administered in the first and second doses (heterologous or homologous). Furthermore, the vaccine effectiveness level ranged from 88.8% to 92.5% during first three months after authorization.9 The infectious agents (related to mutation and immune escape), host-related characteristics (age, comorbidities, immune status, genetics), and vaccine factors (dosing intervals, administration routes, types) contribute to the immune response (as shown by antibody levels) formation after booster vaccination.10,11
Our study discovered a higher percentage of GG genotype (76.9%) than GA genotype (23.1%) as the evidence of TNF-α– 308G/A promoter polymorphism. The same phenomenon was also discovered in prior research in Indonesia; however, the GG prevalence was only 51%. The differences may be attributed to ethnic-specific variation and types of subject.12 This cytokine-related gene has been linked with the progression of several infectious diseases and conditions, most notably influencing susceptibility to pulmonary tuberculosis, predisposing to sepsis, and influencing COVID-19 progression.12 Specifically, people who have the A allele of the TNF-α polymorphism are more vulnerable to COVID-19 infection and are more likely to have severe disease state.7
The impact of cytokine gene polymorphism in COVID-19 cases has been described in terms of susceptibility or outcome following infection.13 However, evaluation of this phenomenon concerning antibody production following COVID-19 vaccination has yet to be reported.
Although not extensively studied in COVID-19 vaccination, several vaccine activities have been linked to host genetic factors. Previous research on the antibody response to inactivated Japanese encephalitis vaccine discovered that TNF-α genes significantly influenced the vaccine's seropositivity. The polymorphism of this gene also had a significant impact on the amount of antibodies produced.14 Earlier research on the response to hepatitis B vaccination revealed the contribution of the HLA-DRB1, HLA-DQB1, and HLA-DPB1 genes in facilitating the outcome of vaccination.15 A broader set of genetic polymorphisms, including HLA, cytokine, innate immune system, and viral receptor gene, explained nearly 30% of the inter-individual variation in measles vaccine-specific humoral immunity.16
There are several limitations in our study. First, the persistence of antibody levels after the third vaccination dose (booster) was not evaluated in this study. It is critical to determine the efficacy of the booster vaccination to improve protection and minimize potential severe disease states. Second, we did not have a complete perspective of IFNL3 (CT) and IFNL4 (TG) gene-related polymorphism since that gene was only established in one subject, associated with a small sample size (this is a pilot study). Third, baseline level of antibody prior to the third vaccination was not determined. It is important to evaluate whether the surge of antibodies are solely related to the vaccine or not.
ConclusionAnti-SARS-CoV-2 spike protein IgG antibody level following the third dose vaccination may be influenced by the polymorphism of the cytokine gene, particularly the one related to TNF-α (GG genotype). A more comprehensive analysis of the observed gene related to immunity on a larger population scale is suggested to improve the evidence. It can be accomplished by future multicenter cohort studies that examine a broader range of factors influencing vaccination-related immunity, such as ethnicity, race, comorbidities, behavioral, and nutritional factors.
Funding sourceThis study was partially funded from Sriwijaya University Faculty of Medicine grant (statute number: 0330/UN9.FK/TU.SK/2021).
Ethical approvalThe Sriwijaya University Faculty of Medicine Ethics Committee granted ethical approval for this study (approval number: 062–2021).