To standardize an RNA (ribonucleic acid) extraction protocol in children's saliva specimens.
MethodsThe study was conducted on saliva specimens from 60 children who participated in the study with their parents’ authorization. Comparison of two RNA extraction methods was established; methods assessed concentration, quality and yield. Moreover, genic expression of GUSB gene was achieved with RT-PCR (reverse transcriptasepolymerase chain reaction). Data were analyzed through measurements of central tendency and dispersion, frequencies and percentages. T-Student and χ2 tests were used in order to differentiate between both methods (p < 0.05).
ResultsAnalysis of cell amounts per saliva ml revealed a mean of 564,977.8 (SD = 246,678.6); a RNA in children's saliva extraction method was standardized, specifically the method using RNeasy® Protect Saliva Mini Kit Qiagen exhibited better characteristics of RNA concentration (p = 0.0000) and yield (p = 0.0000) when compared to the method using QIAzol®; no RNA (p = 0.146) quality differences were found.
ConclusionsRNA could be extracted from children's saliva specimens, therefore, it is suggested to use saliva for the molecular analysis of different oral and systemic diseases.
Estandarizar un protocolo de extracción de ARN en muestras de saliva de niños.
MétodosA partir de muestras de saliva provenientes de 60 niños que participaron previa autorización de sus padres. Se compararon dos métodos para extracción de ARN, evaluando concentración, calidad y rendimiento; además se realizó la expresión génica del gen GUSB a través de la técnica RT-PCR (transcriptasa reversa-reacción en cadena de la polimerasa). Los datos fueron analizados mediante medidas de tendencia central y dispersión, frecuencias y porcentajes, para establecer diferencias se utilizaron las pruebas t-Student y χ2 (p < 0.05).
ResultadosAl analizar la cantidad de células por mL de saliva se encontró una media de 564,977.8 (DE = 246,678.6); se logró estandarizar un método de extracción de ARN en saliva de niños, específicamente el que utilizó RNeasy® Protect Saliva Mini Kit-Qiagen mostró mejores características de concentración de ARN (p = 0.0000) y rendimiento (p = 0.0000) al compararlo con el que usó QIAzol®; no existieron diferencias en la calidad del ARN (p = 0.146).
ConclusiónEl ARN pudo extraerse de muestras salivales de niños, por lo cual se sugiere el uso de la saliva para análisis molecular de diferentes enfermedades sistémicas y de la cavidad bucal.
Use and development of non-invasive techniques for biomarker identification is a promising tool for diagnosis of diseases in the mouth and general systemic health circumstances.1 In this sense, saliva has rapidly emerged as a non-invasive source possessing important biological information.2,3 Saliva is formed after blood filtration into the salivary glands, and contains electrolytes, proteins, microorganisms and genetic material, including deoxyribonucleic acid (DNA) and ribonucleic acid (RNA) in cell portion and free cells.4 As a product of blood filtration, genetic material in saliva's free cells originates from different sources in the body, whereas that coming from cell fraction is almost exclusively derived from the oral mucosa.5
RNA can be in the mouth from different sources, including saliva secretion from major and minor salivary glands, crevicular gingival fluid and desquamation or oral epithelial cells.6 Varied secretions of salivary glands of micro and macro molecules could originate in acinar cells or due to circulation. Frequently another RNA source could be the desquamation process of epithelial cells of the mouth.7–9
Saliva is an important component for processes such as speech and digestion; it is equally a means of protection against microorganisms.10 Additionally, biomarkers for bacterial, viral and fungal infections have been reported in saliva in systemic diseases as well as local conditions.11,12 These markers can come from varied molecular species from proteins and antibodies to DNA and RNA.6 Recently, RNA obtained from free cells in human saliva showed it was suitable to be used for oral cancer biomarkers study,13–15 by means of PCR techniques; it has been likewise suggested that free cells in the saliva of healthy subjects contain more than 3,000 species of mRNA.16 Little is known of RNA properties and molecular nature,17 one of the main problems is the amount of compounds selected in the saliva when compared to blood, another problem is to ability to defferentiate between microbial and human origin transcripts; the limited amount of salivary RNA requires more sensitive and specific methods.18 It has equally been suggested that saliva contains ribonucleases from various sources; this could possibly hinder analysis of RNA in saliva.6 It is still not completely clear how RNA and ribonucleases can coexist in saliva, a possible explanation is that endogenous salivary RNA is protected from degradation just like what occurs with plasma RNA.19,20 Likewise, it has been reported that saliva contains mRNA fractionally degraded chains18 and that RNA degradation in saliva is relatively slow when compared to endogenous RNA;6 this would suggest presence of salivary RNA stabilization mechanisms. Presently, all these obstacles are being subject of study, due to this fact, increased interest in salivary RNA research could be expected.
Among advantages of using saliva specimens as opposed to blood specimens, the following benefits have been pointed out: harvesting method is safe, painless and non traumatic, harvesting technique is easy to achieve, not requiring specially trained personnel, harvesting equipment is simple, and saliva specimens can be taken several times without incurring in patient's discomfort.21,22 Thus it is necessary to standardize molecular biology techniques using saliva study as medium; this could contribute with diagnosis and prognosis of some diseases; it could also contribute to achievement of research in genomic areas, which could help to better understand physiopathology of these diseases from a molecular perspective.
The aim of the present research was to standardize a RNA extraction protocol in saliva specimens harvested from children.
Material and MethodsParticipantsAll 60 children participating in the sample were treated at dental clinics of the University of Cartagena. Average age was 6.8 (SD = 4.6) years. Participants did not report personal history of neoplastic disease, immunodeficiencies, autoimmune disorders or hepatitis. For children to participate, parents had to previously sign a written informed consent form. The study was approved by the ethics committee of the University of Cartagena.
Saliva specimen harvestingNon-stimulated human saliva specimens were collected in polypropylene tubes placed in ice, in amounts of 3 mL. All children were instructed to wash their teeth and not to consume food or drink during the hour preceding saliva specimen collection. All specimens were harvested in the Biochemistry Laboratory of the Cartagena University, following autonomous salivation protocol. They were then stored at -80 oC until further analysis.
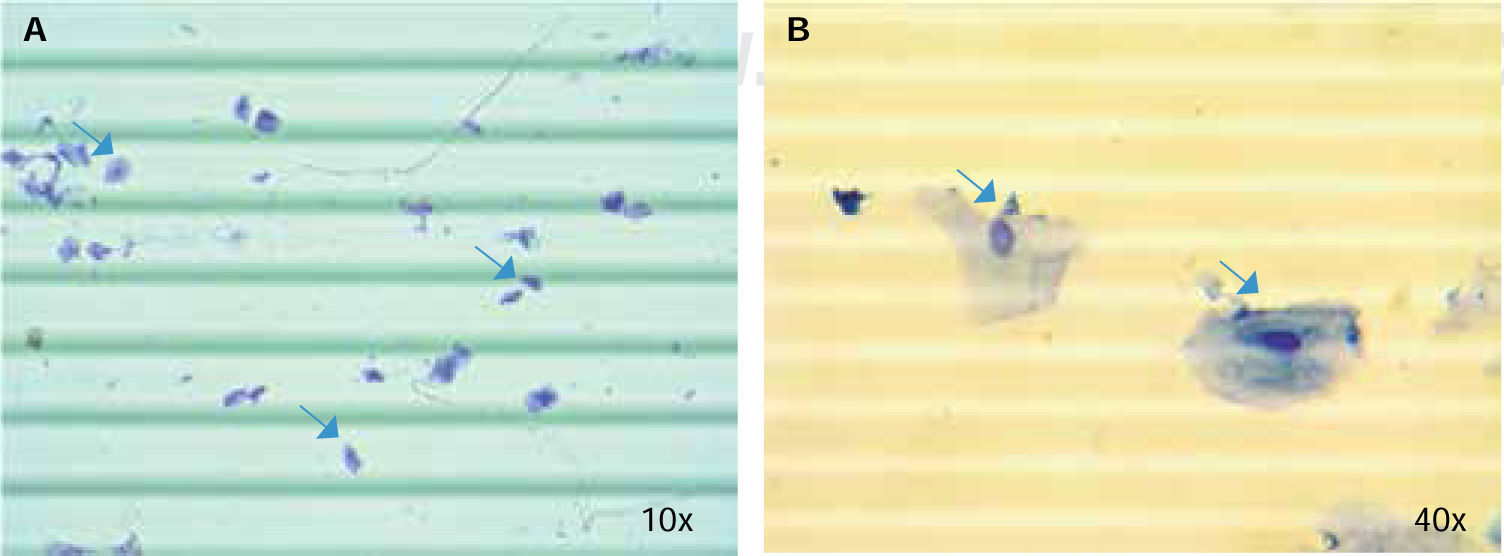
Trypan blue cell countPreviously reported procedures were taken into account for this essay;23 20 μL saliva specimens were mixed with 20 μL trypan blue in a 0.6 mL Eppendorf tube; 10 μL of this new solution were pipetted at each side of a hemocytometre. An optic microscope was then used (Nikon SE) was set at 10x and cells of all four quadrants located at the corners of the Neubauer (BOECO) chamber were quantified. Average cell number was thus obtained, it was then multiplied by diffusion factor and by 10,000 in order to calculate total cell number in 1 mL saliva.
RNA extraction in salivaThe following protocols for RNA extraction in saliva were used and evaluated:
Protocol A: in this method lysis QIAzol® (Qiagen brand) (TRIzol) reactive was used; manufacturers recommendations were followed with some modifications. Initially, 500 μL TRIzol were added to 500 μL saliva specimens in a 2 mL Eppendorf tube, they were then incubated at 25 oC for 10 minutes. After this 100 μL of chloroform:isoamylic (24:1) were added, achieving vortex for 15 seconds; the specimen was incubated at room temperature for five minutes and was centrifuged at 10,000 rpm during 10 minutes at 4 oC. Supernatant was then collected in a new 1.5 mL Eppendorf tube, to which 250 μL isopropanol were added, achieving vortex for 15 seconds. The specimen was incubated overnight at 4 oC, after this, it was centrifuged at 10,000 rpm during 15 min at 4 oC, supernatant was eliminated through inversion, and two washes were applied to the precipitate with 500 μL of 75% ethanol, vortex for 15 seconds and centrifugation at 8,800 rpm for 5 minutes at 4 oC. Supernatant was eliminated by inversion after which the precipitate was dried at room temperature. Finally, precipitate was re-suspended in 20 μL of RNase free water and was stored at 20 oC until further analysis.
Protocol B: RNeasy® Protect Saliva Mini Kit (Qiagen) was used in this project. Manufacturer's instructions were followed with some modifications. Initially, at the moment of harvesting, 200 μL of saliva were taken; 1 mL RNA protect saliva reagent were added to a 2 mL Eppendorf tube, vortex was achieved for 30 seconds, this mixture was then centrifuged for 10 minutes at 12,000 rpm at 25 oC, supernatant was removed using points and micropipettes; 350 μL of Buffer RLT were added into the precipitate, achieving vortex twice for 30 seconds. After this, 350 μL 70% ethanol were added and after pipette homogenization this content was transferred to a RNeasy® MinElute Spin 2 mL column, centrifugation at 10,000 rpm for 15 seconds at 25 oC was achieved, the liquid crossing through the column was discarded; with empty column, centrifugation was conducted at 12,000 rpm for 5 min at 25 oC to this, 350 μL of RW1 buffer were added, centrifuging at 10,000 rpm for 15 seconds at 25 oC. Once again the liquid that passed through the column was discarded and 500 μL of RPE buffer were added, centrifuging then at 10,000 rpm for 15 seconds at 25 oC, the liquid that came across the column was discarded, 500 μL of 80% ethanol were added to it to be then centrifuged at 10,000 rpm for 2 minutes at 25 oC, the liquid that traversed the column was again discarded and a centrifugation as performed. Residual liquid was discarded, collection tube was replaced by a new one, 14 μL of RNase-free water were incorporated into the center of the column's membrane; centrifugation at 10,000 rpm for 1 min at 25 oC was performed, isolated RNA was collected and preserved at -20 oC until further analysis.
Assessed operative characteristicsThe following parameters were assessed for both protocols:
RNA concentration: evaluation of RNA concentration was achieved with spectrophotometry at 230, 260 and 280 nm in NanoDrop 2000 UV-Vis Spectrophotometer (Thermo Scientific). Concentration value was provided by the device. Nevertheless, this concentration value was rectified by multiplying x 40 absorbance at 260 nm. These measurements were conducted twice for each specimen.
RNA quality: in the Cartagena University Immunology Laboratory, obtained purity degree of A260/A280; absorbance relation was considered in the NanoDrop 2000 UV-Vis Spectrophotometer (Thermo Scientific) device. A relation of 1.5 to 2 was considered an indicator of suitable purity degree, therefore, this RNA was in an acceptable quality range.
Yield: was determined bearing in mind relation between saliva RNA concentration and number of cells for each specimen, considering as ng quantity in RNA among number of cells found in 1 mL of saliva.
Genic expression analysis through RT-PCRAt a later point, with the protocol that exhibited better results, total RNA was used to obtain cDNA, which was used for conventional PCR amplification, using primers (5’-ATCACCGTCACCACCAGCGT-3’/3’GTCCCATTCGCCACGACTTTGT-5’) corresponding to the gene of glucoranidase beta (GUSB), dNTPs (10 mM) MgCl2 (25 mM) and one unit of Ecotaq polymerase. PCR reaction was conducted in a thermocycler (BIO-RADT100™) under the following circumstances: 94 o C during 30 seconds, 72 oC for one minute and a final elongation time at 72 oC during 10 minutes.
PCR products were visualized through electrophoresis in agarose gels at 1.0%, in TAE tampon 1x (Tris 40 mM; 0.1% glacial acetic acid; EDTA 1 mM); containing EZ-Vision® (Amresco®) and UV light exposition in the device ChemiDoc™ XRS+ System. All specimens were analyzed three times.
Statistical analysisStatistical analysis was initiated through the design of an Excel Microsoft® Office 2010 database, which was then transported to the program STATA® (StataCorp LP, College Station TX USA). Initially data normalcy assumption was performed based on Shapiro Wilk test. Measures of central tendency, dispersion and proportions were used for the descriptive analysis. T-Student test for quantitative variables was used for protocol comparison, and χ2 test was used for qualitative variables. In all test assumption was made of 0.05 decision limit probability.
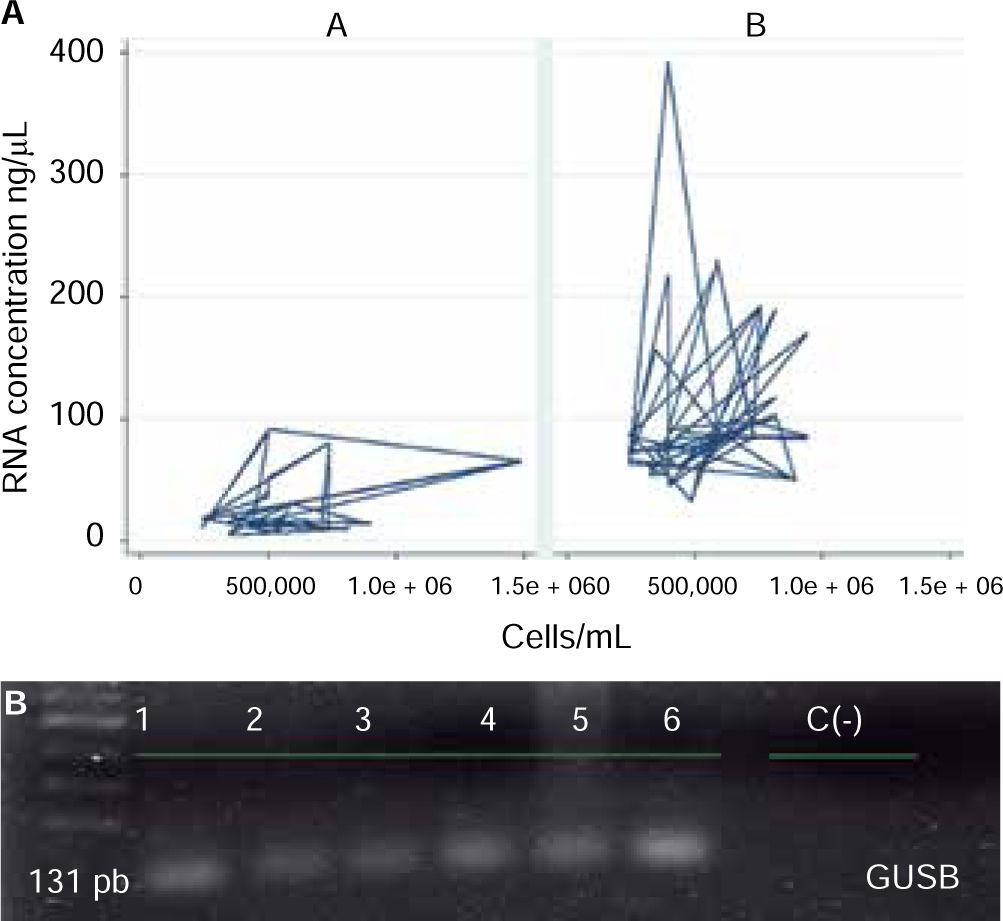
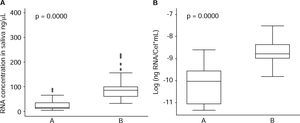
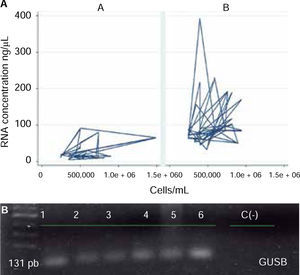
ResultsAnalysis of cell amount per saliva ml revealed a mean of 564,977.8 (SD = 246,678.6), with range varying from minimum value of 242,500 cell/mL and maximum value of 1,487,500 cell/mL (Figure 1).
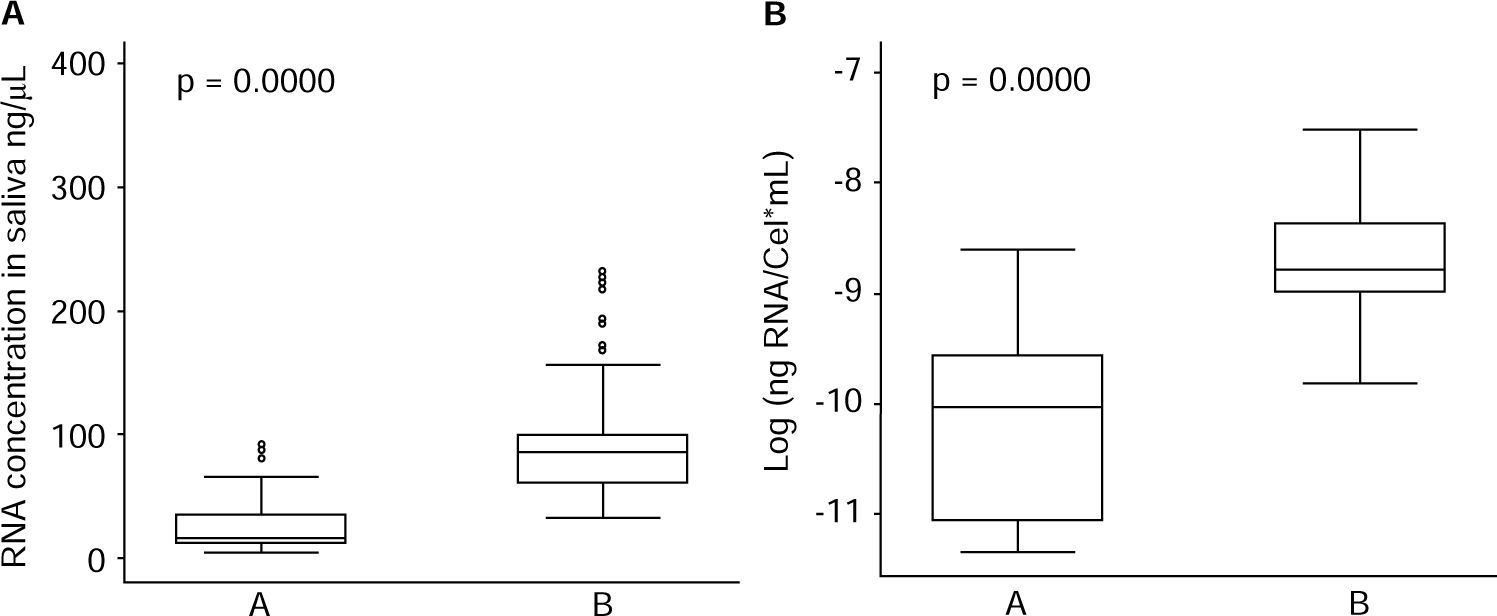
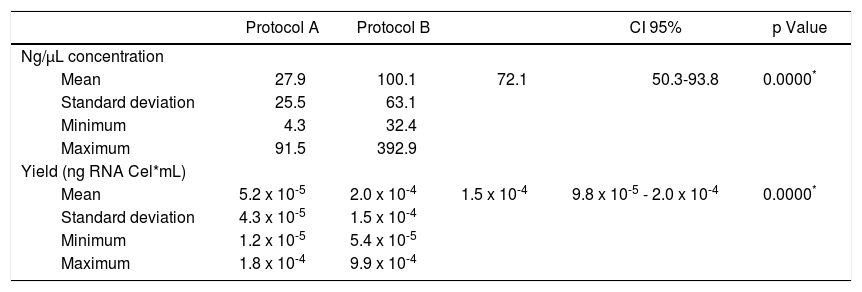
With respect to operative characteristics of both assessed protocols for RNA harvesting in saliva, it was found that global concentration mean of isolated RNA in saliva specimens was 71.0 (SD = 62.4) ng/μL, where minimum value was 4.3 ng/μL and maximum value was 392.9 ng/μL. For protocol A (QIAzol®), concentration mean was 27.9 (SD = 25.5) ng/μL; for protocol B (RNeasy® Protect Saliva Mini Kit Qiagen) it was 100.1 (SD = 63.1) ng/μL, as far as yield was concerned, global mean was 1.4 x 10-4 (SD 1.3 x 10-4) ng RNA/Cel*mL, with minimum value of 1.2 x 10-5 ng RNA/Cel*mL and maximum value of 9.9 x 10-4 ng RNA/Cel*mL. For protocol A it was 5.2 x 10-5 (SD 4.3 x 10-5) ng RNA/Cel*mL; for protocol B it was 2.0 x 10-4 (SD = 1.5 x 10-4) ng RNA/Cel*mL (Table I). Comparison of RNA harvesting methods in saliva specimens revealed statistically significant differences with respect to RNA concentration (p = 0.0000) and yield (p = 0.000) (Figure 2), where protocol B exhibited more favorable results than protocol A.
Comparison of protocols for RNA extraction in saliva.
| Protocol A | Protocol B | CI 95% | p Value | ||
|---|---|---|---|---|---|
| Ng/μL concentration | |||||
| Mean | 27.9 | 100.1 | 72.1 | 50.3-93.8 | 0.0000* |
| Standard deviation | 25.5 | 63.1 | |||
| Minimum | 4.3 | 32.4 | |||
| Maximum | 91.5 | 392.9 | |||
| Yield (ng RNA Cel*mL) | |||||
| Mean | 5.2 x 10-5 | 2.0 x 10-4 | 1.5 x 10-4 | 9.8 x 10-5 - 2.0 x 10-4 | 0.0000* |
| Standard deviation | 4.3 x 10-5 | 1.5 x 10-4 | |||
| Minimum | 1.2 x 10-5 | 5.4 x 10-5 | |||
| Maximum | 1.8 x 10-4 | 9.9 x 10-4 |
Protocol A (QIAzol®) and B (RNeasy® Protect Saliva Mini Kit-Qiagen).
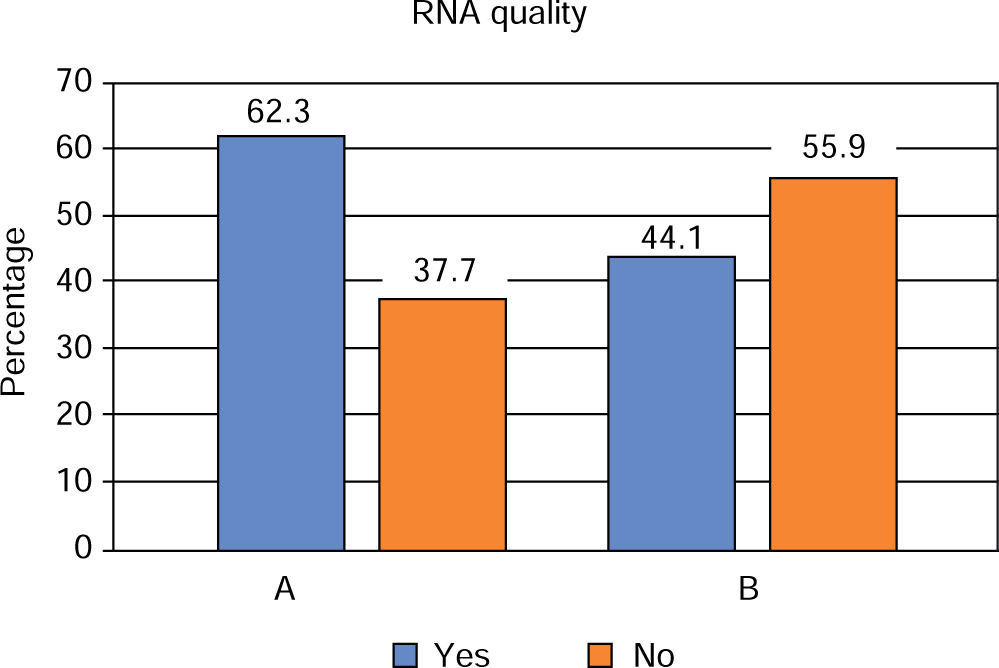
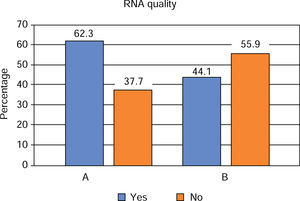
With respect to quality of RNA harvested in saliva, it was observed that 41.1% was found to be in the optimum ranks of 1.5 to 2.0 (A260/280); protocol A met in 62.3% with this condition and protocol B in 44.1%. Comparison of this characteristic with protocol type did not reveal statistically significant differences between them (p = 0.146) (Figure 3).
Evaluation of protocol behavior according to operative characteristics revealed that protocol B showed better behavior than protocol A, since, in general terms, better concentrations or RNA in ng/μL were obtained using lesser amounts of cells/mL saliva. Likewise, this protocol exhibited genic expression of GUSB normalizing gene (Figure 4).
DiscussionA standardization method was achieved when dealing with RNA extraction procedures in saliva specimens; this would suggest that saliva is an excellent method for molecular biology studies; this concurs with Park6 who characterized salivary RNA. Likewise Li16 had previously reported presence of human mRNA in saliva specimens. Additionally Chiang24 recognized that stabilization and processing of transcriptome represents a critical challenge in the study of salivary biomarker, due to the ubiquitous nature of nucleases and proteases, as well as the inherent instability of these biomarkers. Therefore, this standardization is considered and advance and a contribution to research, it will promote other research projects which will evaluate salivary biomarkers as well as molecular mechanisms associated to systemic and oral diseases.
Results suggest that the method using RNeasy® Protect Saliva Mini Kit-Qiagen showed better RNA concentration and yield characteristics when compared to the method using QIAzol®. This could be explained bearing in mind that RNeasy® protect saliva Mini Kit-Qiagen contains a protecting solution which stabilizes salivary RNA, Fabryova18 considers it represents the best salivary RNA stabilizing reactive that is presently available; this was determined through compassion of Ct values in RNA specimens treated with stabilizing solutions and stored at room temperature for 10 days.25 Usage of these stabilizing solutions is the first fundamental step for the later analysis for saliva RNA extraction, due to the fact that there are reports on the existence of salivary ribonucleases which remain active after saliva harvesting, which would in turn justify its inhibition before processing those specimens.18 Another possible explanation would be that this method uses poly A carrier among its reagents; this reagent contains an RNA bonding protein which bonds to the poly extremity (A) of the mRNA, located at the 3’ extreme, this is a widely known RNA protection mechanism. It has been suggested that in eukaryotes many proteins bond directly or indirectly to RNA to achieve its stabilization. Over 30% of mRNA found in human saliva contains areas rich in adenine and uridine which increases RNA stability, this proportion is found five-fold when compared to that observed in other mRNA of the human body.26 Likewise, areas rich in A and T at the 3’ extreme stabilize it before translation; bearing these mechanisms in mind, Khabar27 states that average salivary mRNA preserves 42% of its original longitude.
On the other hand, Pandit 28 also compared two methods of RNA extraction in saliva, likewise using one method with QIAzol® and the other with a commercial kit. He stated that the QIAzol® method produced a high total RNA yield from saliva, showing suitable absorbance relation measured at 260 and 280 nm; he also stated that the commercial kit produced a ten times lesser RNA yield. Therefore, he suggested use of QIAzol® lysis reagent to isolate RNA specimens of saliva specimens stores without RNAse inhibitors at -80 oC during more than two years. Likewise, Detz4 compared Qiagen RNA Protect® Saliva Mini Kit and QIAamp Viral RNA Mini kit for extraction of RNA of free cells and supernatant of neonatal saliva. He concluded that although in mbopth methods mRNA was extracted and amplified from all saliva supernantant specimens, the method of QIAamp Viral RNA Mini kit showed better results with respect to RNA amount and concentration. In this sense, Maron29 compared RNA yield, quality stability and yield of RTqPCR in the systems Qiagen RNeasy® Protect Saliva Mini kit and DNA Genotek Oragene-RNA® in the saliva of newborns. He suggested that although the Qiagen essay can decrease general extraction time, RNA yield and yield in later transcriptomic analysis is more robust when using Genotek DNA essay; he nevertheless clarified that RNA integrity did not differ among these methods.
It should be pointed out that a possible explanation for these differences might be due to circumstances inherent to each experiment, operator and laboratory, therefore it is recommended to use the standardized method described in this study, bearing in mind characteristics and circumstances of where the method is going to be applied. In this sense, Grabmüller compared five kits readily available in the market for RNA extraction (mirVana™ isolation kit miRNA Ambion, Trizo® reagents, Invitrogen, NucleoSpin® miRNA kit Macherey-Nagel, AllPrep DNA/RNA Mini Kit and RNeasy® Mini Kit, both from Qiagen) in order to assess their relative effectiveness to produce good quality RNA using specimens of small amounts of cells, among which were saliva and oral mucosa. He suggested that although there are considerable differences among yields of quality values for RNA and expression values, in general, there was not one best method to satisfy all demands established by different RNA and DNA analyses, therefore, it so seemed that each method exhibited specific advantages and disadvantages, thus, he recommended to carefully choose among available methods and adjust their characteristics with the requirements of available experimental environment. In a similar manner, Sellin31 compared yield of five commercially available kits for total RNA extraction from specimens with small amounts of cells; he concluded that each kit was generally able to extract required RNA amount for genic expression applications or other essays. Nevertheless, differences in quality of RNA extracted through all of the kits indicate that they can differ in their capacity to produce acceptable RNA in some applications; this would suggest there are practical differences among commercially available RNA extraction kits, and that they must be taken into account when selecting extraction methods to be used in RNA isolation which in turn will lead to genic expression analysis.
In this sense, through the standardized method, in the present study we achieved analysis of genic expression of GUSB normalizing gene, which suggests the effectiveness of RNA extraction method and the possibility to use saliva as a study specimen for different biomarkers in child population. This entails many benefits, among which we can count ease of harvesting, and painless and non-invasive method. Likewise, Li13 when dealing with saliva specimens of healthy subjects, achieved identification of genic expression of 185 transcript types, which were found in each individual. It has likewise been reported that transcripts of B-actin, RPS9, glyceraldehyde-3-phosphate dehydrogenase (GAPDG), IL8, sperdimine/spermine and N1 acetyltransferase are present in high concentrations in saliva specimens.16
Briefly, salivary RNA is a promising tool since it can provide information not only of gene presence, but also of gene expression. This could be explained by the fact that saliva is a plasma ultrafiltration, which indicates it could also be used for diagnosis of conditions unrelated to the oral cavity.18 In this sense, saliva can be considered as a viable means for molecular studies, since a RNA extraction method in children's saliva could be achieved, specifically, the method using RNeasy® Protect Saliva Mini Kit-Qiagen showed better RNA concentration and yield characteristics, always considering inherent properties of operator and laboratory, therefore its use is suggested for studies on population's genomic and biochemical signaling, which might contribute to the understanding of molecular mechanisms, as well as the search of salivary biomarkers of different systemic and oral diseases.
DDS, Degree in Biochemistry. Degree in Clinical Epidemiology, Frontera University. Associate Researcher in Group GISPOUC, School of Dentistry
This article can be read in its full version in the following page: http://www.medigraphic.com/facultadodontologiaunam
Director of Group Prometeus & Biomedicine, Applied to Clinical Sciences. Pharmaceutical Chemist, PhD in Biochemistry and Molecular Biology, Complutense University, Madrid. Teacher at the School of Medicine