The Saccharomyces cerevisiae vacuole is actively involved in the mechanism of autophagy and is important in homeostasis, degradation, turnover, detoxification and protection under stressful conditions. In contrast, vacuolar proteases have not been fully studied in phylogenetically related Candida glabrata.
AimsThe present paper is the first report on proteolytic activity in the C. glabrata vacuole.
MethodsBiochemical studies in C. glabrata have highlighted the presence of different kinds of intracellular proteolytic activity: acid aspartyl proteinase (PrA) acts on substrates such as albumin and denatured acid hemoglobin, neutral serine protease (PrB) on collagen-type hide powder azure, and serine carboxypeptidase (CpY) on N-benzoyl-tyr-pNA.
ResultsOur results showed a subcellular fraction with highly specific enzymatic activity for these three proteases, which allowed to confirm its vacuolar location. Expression analyses were performed in the genes CgPEP4 (CgAPR1), CgPRB1 and CgCPY1 (CgPRC), coding for vacuolar aspartic protease A, neutral protease B and carboxypeptidase Y, respectively. The results show a differential regulation of protease expression depending on the nitrogen source.
ConclusionsThe proteases encoded by genes CgPEP4, CgPRB1 and CgCPY1 from C. glabrata could participate in the process of autophagy and survival of this opportunistic pathogen.
La vacuola de Saccharomyces cerevisiae está involucrada activamente en el mecanismo de autofagia, desarrollando una labor importante en la homeostasis, degradación, recambio proteico, desintoxicación y protección de la célula en condiciones de estrés. Por el contrario, las proteasas vacuolares de Candida glabrata aún no han sido estudiadas por completo.
ObjetivosEl presente trabajo describe por primera vez la actividad proteolítica vacuolar en C. glabrata.
MétodosLos estudios bioquímicos realizados en C. glabrata pusieron de manifiesto la presencia de diferentes actividades proteolíticas: aspartil proteinasa ácida, que actúa sobre sustratos como la albúmina y la hemoglobina ácida desnaturalizada; serín proteasa neutra, con actividad sobre el substrato de tipo colágeno hide powder azure, y serín carboxipeptidasa, que actúa sobre N-benzoil-tyr-pNa.
ResultadosLa obtención de una fracción subcelular mostró una elevada actividad enzimática específica de las tres proteasas, lo que permitió confirmar su localización vacuolar. Se realizaron análisis de la expresión de los genes CgPEP4 (CgAPR1), CgPRB1 y CgCPY1 (CgPRC1), codificantes de las actividades proteolíticas aspartil proteasa A, proteasa neutra B y carboxipeptidasa Y, respectivamente. Los resultados reflejan una regulación diferencial de la expresión de la proteasa, dependiendo de la fuente de nitrógeno.
ConclusionesLas proteasas codificadas por los genes CgPEP4, CgPRB1 y CgCPY1 podrían participar en el proceso de autofagia y supervivencia de este patógeno oportunista.
The fungal vacuole is a dynamic and acidic compartment of the cell, similar to lysosomes, with a wide variety of hydrolytic enzymes such as proteases, trehalase, alpha-mannosidase, and alkaline phosphatase. The main functions of these enzymes are the degradation of proteins, storage and bidirectional transport of ions and metabolites, detoxification, ion homeostasis, and maintenance of cytosolic pH.15,19,23,30,33 The Saccharomyces cerevisiae vacuole is an important organelle for the adaptation to new environments. Its morphology changes readily for the purpose of survival in response to extracellular signals, as well as signals related to stress conditions and cell differentiation.2,19,40,44 In Candida albicans, the vacuole allows the activation of certain virulence factors, such as differentiation of the yeast-hyphae, as well as adhesion and survival within macrophages.17,27,29
Candida glabrata, an opportunistic yeast pathogen that causes candidiasis, has an unknown sexual cycle. The C. glabrata haploid genome is phylogenetically more related to S. cerevisiae, with approximately 12.3 Mb organized in 13 chromosomes.5,7,8,31 In order to survive, proliferate and evade host defense strategies, this yeast must be able to adapt to changes in the microenvironment of the host during invasion and systemic spreading; however, the underlying mechanisms are not well understood.32,34 The gene expression of some extracellular aspartyl proteases of C. glabrata has only been studied in a mouse model and macrophages.14 The vacuolar protease gene expression under different physiological conditions (including nutrition) has not yet been reported. In this study, we describe the biochemical activity and regulation of various vacuolar proteases of C. glabrata in order to infer the potential functional role of each of these enzymes.
Materials and methodsStrains and growth conditionsThe CBS138 strain of C. glabrata used in this study was kindly provided by Dr. Bernard Dujon from Institut Pasteur, France.7 The yeast strain was stored at −70°C in 30% glycerol. Cells were grown in YPD medium (1% yeast extract, 2% peptone, and 2% glucose) at 37°C for 48h with constant shaking, and then inoculated into fresh YPD medium.
Culture media used to measure expression of putative genes coding for proteasesSeveral media were tested to determine the effects on C. glabrata genes coding for protease expression. The C. glabrata cells were harvested from YPD medium during early stationary phase (15h) and then washed twice with minimal medium consisting of 0.17% yeast nitrogen base (YNB) without amino acids or ammonium sulfate. The nonproliferating cultures were incubated at 37°C for 6h with constant shaking in YPD medium and in 0.17% YNB supplemented with 2% glucose (YNBg) under five different nitrogen sources (either no nitrogen source, 0.5% ammonium sulfate, 0.2% bovine serum albumin (BSA), 2% proline, 2% peptone).
Preparation of crude extractsCultures in early stationary phase were harvested by centrifugation at 5000×g at 4°C for 10min. Biomass cells were fragmented in a FAST Prep-24 using glass beads (7.5g of glass beads, 12ml of 0.1M Tris–HCl at pH 7.5, and 5g of cells) and pulses (3×20s at 6.5m/s with 2min intervals on ice). Broken cells were centrifuged at 5000×g at 4°C for 10min. The crude extract was carefully removed from the glass beads and centrifuged at 23,000×g at 4°C for 10min. The supernatant was removed and centrifuged at 100,000×g at 4°C for 1.5h using a Beckman ultracentrifuge. The corresponding soluble fraction or cell free extract and membrane fraction were used for enzyme assays and protein determination.
Isolation of intact vacuoles by density gradientIsolation of intact vacuoles was carried out by modifying previously described procedures.26,33,41 Briefly, cells were harvested at early stationary phase of incubation by centrifugation at 2800×g for 5min. They were then washed and resuspended in SOB buffer [1.2M sorbitol, 5mM dithiothreitol (DTT), 50mM Tris pH 7.6] containing Zymolyase 20T (2mgg−1cells). For the liberation of vacuoles after 2h of digestion at 30°C, the spheroplasts were pelleted by centrifugation (3000×g for 10min), resuspended in lysis buffer (1.1M glycerol, 50mM Tris at pH 7.6, and 1mM DTT), and disrupted by a Potter Elvehjem homogenizer in an ice bath. Unbroken cells were eliminated by centrifugation at 3000×g for 10min, and the supernatant was centrifuged at 54,000×g for 30min. The vacuolar pellet was resuspended until a final concentration of 2% sucrose was reached using 15% sucrose and sucrose buffer (10mM Tris pH 7.6, 50mM glycerol, 125mM KCl, 1mM DTT) in order to form a density gradient with sucrose at 15%, 35% and 45% using the same buffer. The sample was then centrifuged at 54,000×g for 40min. The interface (between 15% and 35%) containing the vacuoles was resuspended in solution E (0.6M sorbitol, 5mM Tris at pH 7.6, and 1mM DTT) to a 1:4 proportion and stored at −80°C.
Fluorescence microscopy (FM)When cells were resuspended in SOB buffer, vacuoles were stained adding two markers of the Yeast Vacuole Marker Sampler Kit (Molecular Probes; Life technology, USA). Cell Tracker blue CMAC (7-amino-4-chloromethyl-coumarin) was used to selectively stain the yeast vacuole lumen and a green fluorescent marker (MDY-64) to stain the yeast vacuole membrane. The markers were observed in a fluorescent microscope Imager.M2 (Zeiss, Germany) coupled to the Axio Vision program.
Vacuolar marker enzyme assaysEnzymatic activity was determined using previously described methods.11,12 The following substrates were used for the different proteases: denatured acid hemoglobin for proteinase A activity (PrA), hide powder azure (HPA) for proteinase B activity (PrB), and N-benzoyl-tyrosine-4-nitroanilide (N-BZ-Tyr-4-NA) for carboxypeptidase activity (CpY).22 Protein determinations were performed based on the Lowry method.21
Effect of protease inhibitorsThe effect of inhibitors on protease activity was measured by previously described methods.22 The inhibitors bestatin (100 and 250μM), leupeptin (20 and 50μM), pefabloc (1 and 5μM), pepstatin (2 and 5μM), E-64 (10 and 50μM), 1–10 phenanthroline (1 and 10mM), PMSF (1 and 5mM), and EDTA (1 and 10mM), were employed in this study.
Bioinformatics analysis of canonical vacuolar proteasesThe sequences of putative genes encoding vacuolar proteases of C. glabrata were identified by BLAST analysis (http://blast.ncbi.nlm.nih.gov and http://www.ensembl.org/Multi/blastview). The putative orthologous genes from S. cerevisiae were used as templates in the C. glabrata genome project, available at http://ncbi.labri.fr/Genolevures/elt/CAGL. Promoter sequences were analyzed using the MatInspector database (http://www.genomatix.de/). Protein sequences were deduced from genes using the alternative yeast nuclear genetic code (available at http://www.uniprot.org). These databases contain protein sequences and functional information, as well as cross-references with other databases that display motifs or subcellular location predictions (http://merops.sanger.ac.uk). PCR oligonucleotides were designed by the Primer3 software (http://bioinfo.ut.ee/primer3-0.4.0).
RNA extractionAfter 6h of exposure to the respective media as described above, cells were harvested by centrifugation and frozen immediately. Frozen pellets were washed in diethylpyrocarbonate (DEPC)-treated water and resuspended in RNA isolation buffer (2% Triton, 1% SDS, 10mM NaCl, 1mM EDTA, and 10mM Tris at pH 8.0 in DEPC water). Glass beads were added to constitute 2/3 of the total volume and total RNA was extracted by shaking (4×10s). RNA extraction was performed using the acid phenol method. DNA was removed by treating the RNA with DNase I (Invitrogen, CA, USA), according to the manufacturer's instructions.
Primers and RT-PCRTo amplify fragments of C. glabrata genes encoding vacuolar proteases, specific primer pairs were designed using the gene sequences available at the C. glabrata genome database (http://ncbi.labri.fr/Genolevures/elt/CAGL): CgPEP4 (F: 5′ 691-TATCTGAAGAGTGTC AATGACCCAGC-716 3′, R: 5′ 1208-TACAGCCTCAGCTAAACTGACAACATTGG-1236 3′); Cg PRB1 (F: 5′ 30-AAGAAAGGCGTCTCTCCGGAGG-53 3′, R: 5′ 547-CGTA GTGTTTGGAAGCGATGGTACC-571 3′); and CgCPY1 (F: 5′ 334-ATGACCC TGTCA TCCTATGGTTGAACGG-401 3′, R: 5′ 841-ACTGGGTCAGCACCACTTCCTTCACC-866 3′). The RT reaction for cDNA synthesis was performed using 18S rRNA primers: Cg18S rDNA (F: 5′ 543-CAATTGGAGGGCAAGTCTGG-564 3′, R: 5′ 1267-TAAGAA CGGCCATGCACCAC-1286 3′). The reaction mixture was brought up to a final volume of 20μl, containing 2μg of RNA and 100U Superscript II Reverse Transcriptase (Invitrogen). RT-PCR determinations were performed in duplicate using the reaction conditions described by Loaiza-Loeza.20 Gene expression was normalized according to levels of 18S rRNA expression. The intensity of each amplified band was measured with SigmaGel v.1 (Jandel Scientific Software USA) to obtain the CgProtease/18S rRNA ratio. The standard deviation of each treatment was estimated.
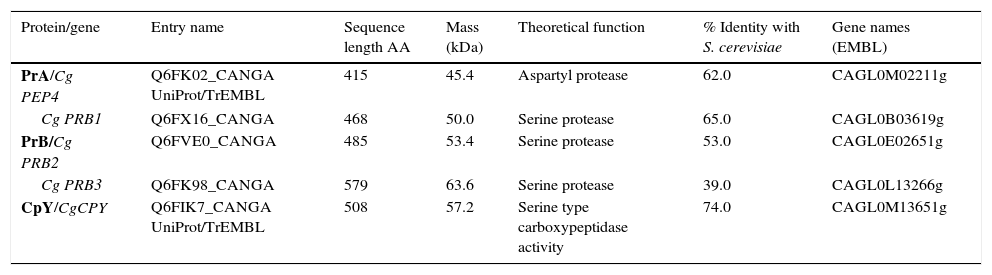
ResultsSequence analysis of C. glabrata putative genes encoding vacuolar proteasesA BLAST analysis based on the C. glabrata genome revealed the presence of five genes likely to encode vacuolar proteases, orthologous to S. cerevisiae genes: CgPEP4 gene coding for aspartyl protease A (PrA), three CgPRB ORFs coding for serine protease (PrB) in three different locus (Cg PRB1, Cg PRB2 and Cg PRB3), and the CgCPY1 gene coding for carboxypeptidase Y (CpY) (Table 1). The bioinformatics analysis suggest that the vacuole could be a putative site of subcellular location of the five predicted proteins, with a certainty factor of 0.9. The deduced proteins from the Cg PEP4, Cg PRB1-3, and CgCPY1 genes showed a high percentage of similarity with deduced proteins from orthologous genes of S. cerevisiae. The molecular masses (Mr) ranged from 45.4 to 63.6kDa (Table 1).
In silico analysis of vacuolar proteases from Candida glabrata.
| Protein/gene | Entry name | Sequence length AA | Mass (kDa) | Theoretical function | % Identity with S. cerevisiae | Gene names (EMBL) |
|---|---|---|---|---|---|---|
| PrA/Cg PEP4 | Q6FK02_CANGA UniProt/TrEMBL | 415 | 45.4 | Aspartyl protease | 62.0 | CAGL0M02211g |
| Cg PRB1 | Q6FX16_CANGA | 468 | 50.0 | Serine protease | 65.0 | CAGL0B03619g |
| PrB/Cg PRB2 | Q6FVE0_CANGA | 485 | 53.4 | Serine protease | 53.0 | CAGL0E02651g |
| Cg PRB3 | Q6FK98_CANGA | 579 | 63.6 | Serine protease | 39.0 | CAGL0L13266g |
| CpY/CgCPY | Q6FIK7_CANGA UniProt/TrEMBL | 508 | 57.2 | Serine type carboxypeptidase activity | 74.0 | CAGL0M13651g |
PrA=Proteinase A; PrB=Proteinase B and CpY=Carboxypeptidase Y.
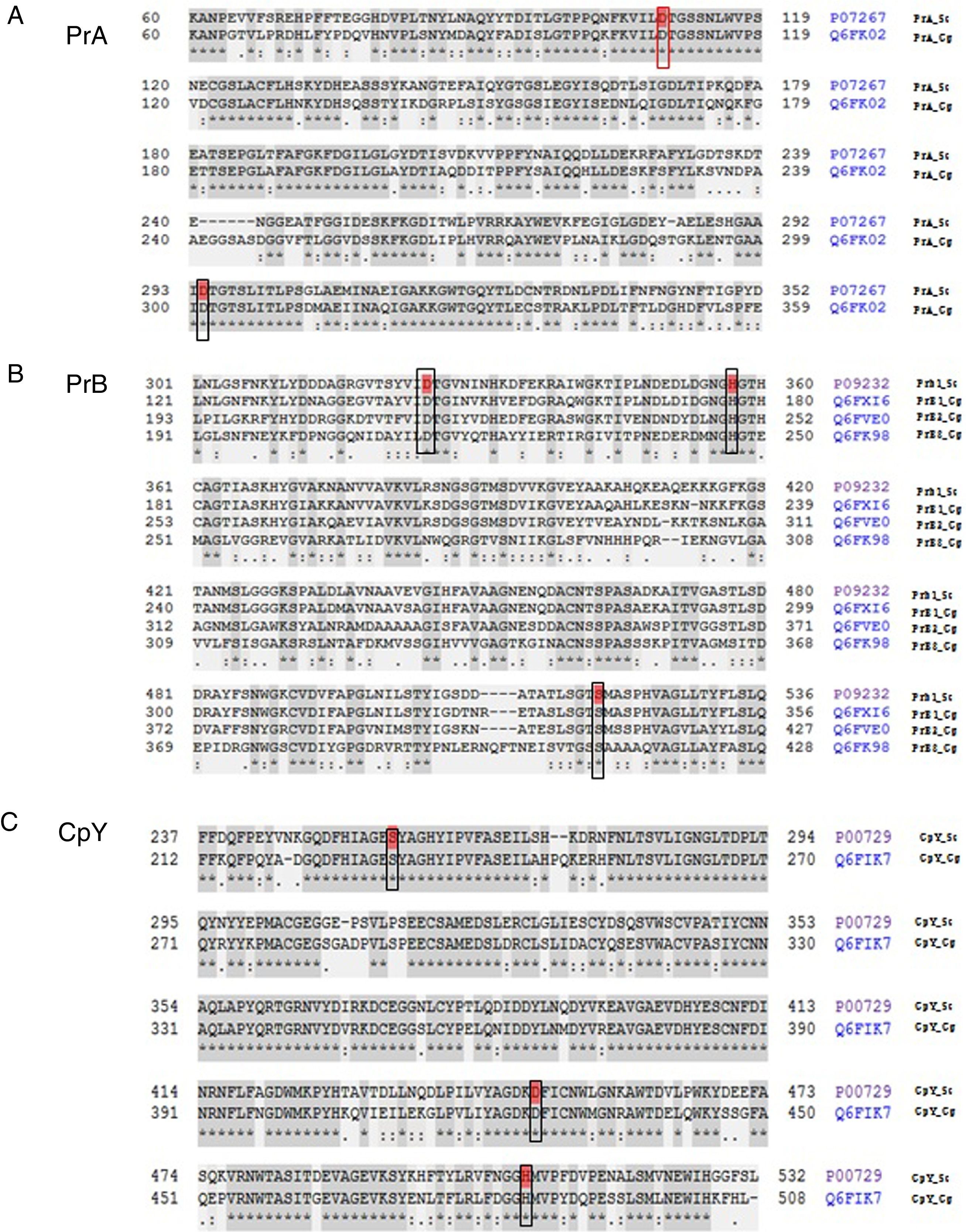
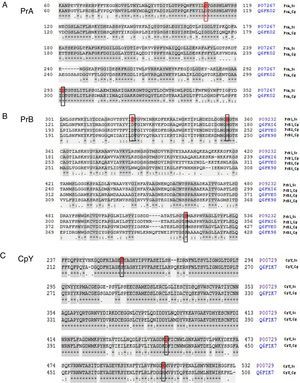
The analysis of the active site of the PrA protein of C. glabrata encoded by the CgPEP4 gene displayed the catalytic domain DD (Asp and Asp), like S. cerevisiae PrA. This catalytic domain is characteristic of aspartyl proteases (Asp109 and Asp294 in S. cerevisiae, and Asp109 and Asp301 in C. glabrata) (Fig. 1).
Peptide sequence alignment of vacuolar proteases. (A) Proteinase A of S. cerevisiae and of C. glabrata in the active DD frame site (Asp109 and Asp294 in S. cerevisiae; Asp109 and Asp301 in C. glabrata). (B) Proteinase B of S. cerevisiae and three putative sequences of C. glabrata. These four sequences show the DHS catalytic triad (Asp325, His357, and Ser519 in S. cerevisiae; Asp145, His177, and Ser339 in PrB1 of C. glabrata; Asp217, His249, and Ser410 in PrB2 of C. glabrata; Asp215, His247, and Ser411 in PrB3 of C. glabrata). (C) Carboxypeptidase in S. cerevisiae and C. glabrata identified in the SDH catalytic triad framed: Ser257, Asp449, and His508 in S. cerevisiae; Ser231, Asp426, and His485 in C. glabrata (http://www.uniprot.org/).
The deduced amino acid sequence alignment of ScPRB1 and CgPRB1-3 shows the serine protease DHS catalytic triad, which in S. cerevisiae is Asp325, His357 and Ser519. In C. glabrata this triad is distributed in different positions in the amino acid sequence of the three proteases PrB [PrB1 (Asp145, His177 and Ser339), PrB2 (Asp217, His249 and Ser410) and PrB3 (Asp215, His247 and Ser411)] (Fig. 1). On the other hand, the carboxypeptidase in S. cerevisiae and C. glabrata showed the serine protease SDH catalytic triad, which in S. cerevisiae is Ser257, Asp449 and His508, and in C. glabrata is Ser231, Asp426 and His485 (Fig. 1).
In order to predict whether the three CgPRB1-3 genes could be differentially expressed, an analysis of their promoter regions (4000pb) was performed. These were compared to the promoter region of PRB1 of S. cerevisiae. Differences were observed in the type and number of the promoter sequences for each PRB genes; likewise, promoter sequences that activate the regulatory genes of nitrogen YNIT (classification according to the nomenclature of the database, MatInspector) were identified in each of the three PRB genes. The CgPRB1 and CgPRB2 genes showed promoter sequences (GATA and URS1, respectively) for the regulation/repression of multiple metabolic pathways of nitrogen. Interestingly, CgPRB1 presented 119 promoter sequences related to the synthesis of amino acids, the regulation of the cellular cycle, resistance to antibiotics, the regulation of pH response, heat shock, and the activation of multi-stress and oxidative stress. Meanwhile, CgPRB2 only showed 6 sequences related to an ARS sequence and transcription factors, and one related to the regulation of metals. Moreover, CgPRB3 has 57 promoter sequences, some of which are shared with the CgPRB1 gene.
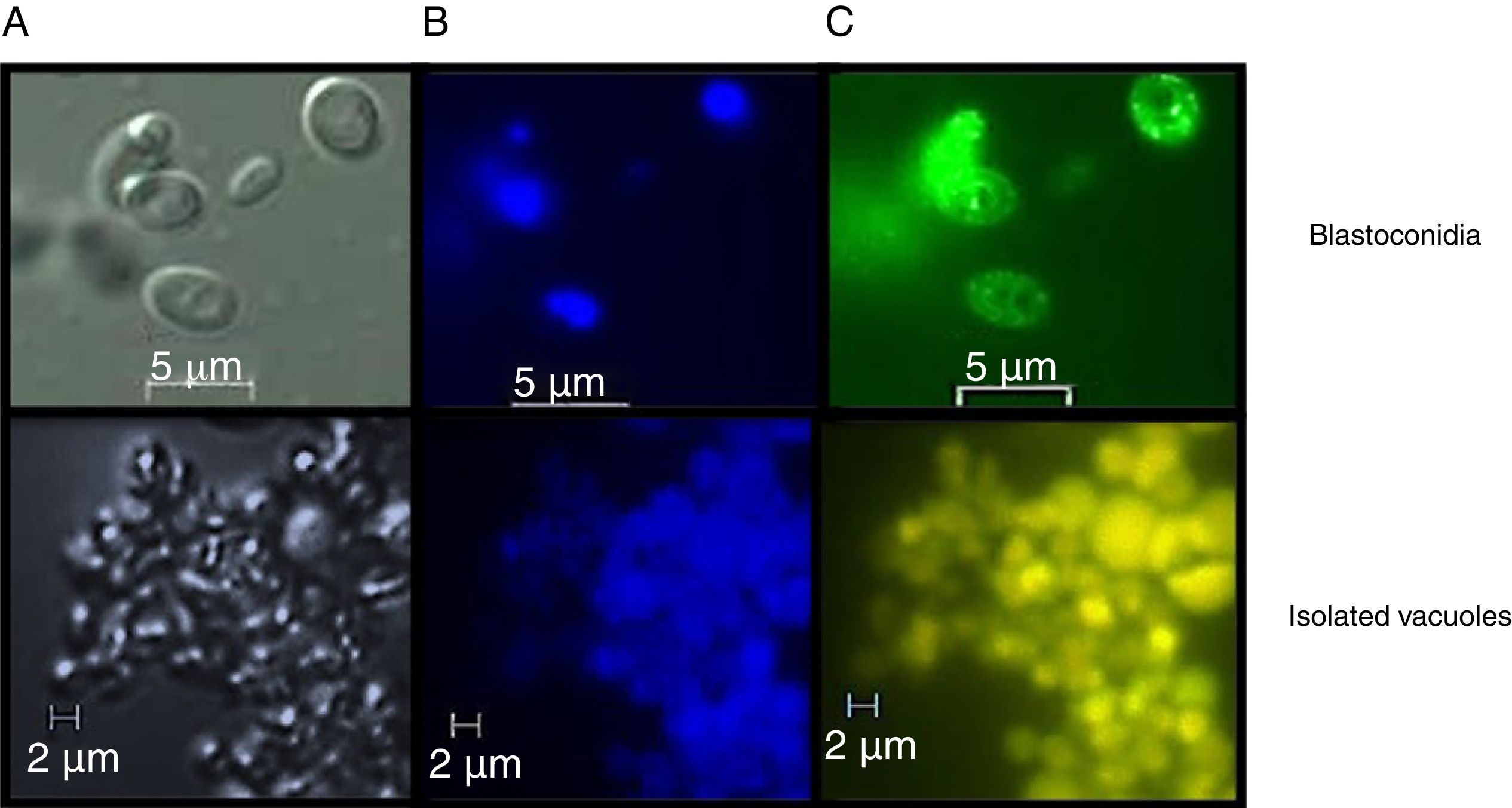
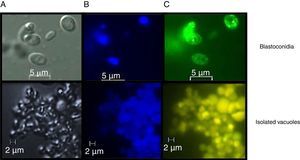
Assessment of vacuolar purity and integrityIntact vacuoles from C. glabrata CBS138 were purified using the procedure described in the Material and Methods section. Fig. 2 shows the location of the vacuoles stained with MDY-64 and CMAC within the C. glabrata blastoconidia. The micrographs obtained by FM confirmed the vacuole integrity after the purification process and showed vacuoles clusters of irregular size.
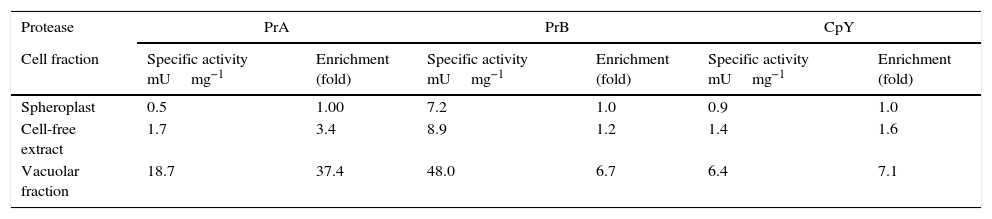
The enzymatic activity, location and inhibition of PrA, PrB, and CpYIn C. glabrata, intracellular activity of PrA, PrB, and CpY was found. These different kinds of proteolytic activity were evidenced during the early stationary phase in YPD medium (15h) (data not shown). These proteases from C. glabrata were found in the soluble (cell-free extract) and vacuolar fraction (Table 2). The enzymatic activity of PrA, PrB and CpY from purified vacuoles yielded enrichment factors of 37, 6.7- and 7-fold, respectively, compared to spheroplasts (Table 2).
Enrichment of the specific activity of vacuolar marker enzymes in intact vacuoles isolated from C. glabrata.
| Protease | PrA | PrB | CpY | |||
|---|---|---|---|---|---|---|
| Cell fraction | Specific activity mUmg−1 | Enrichment (fold) | Specific activity mUmg−1 | Enrichment (fold) | Specific activity mUmg−1 | Enrichment (fold) |
| Spheroplast | 0.5 | 1.00 | 7.2 | 1.0 | 0.9 | 1.0 |
| Cell-free extract | 1.7 | 3.4 | 8.9 | 1.2 | 1.4 | 1.6 |
| Vacuolar fraction | 18.7 | 37.4 | 48.0 | 6.7 | 6.4 | 7.1 |
PrA=Proteinase A; PrB=Proteinase B and CpY=Carboxypeptidase Y.
To establish the proteases types, eight protease inhibitors were applied to the vacuolar fraction from C. glabrata. Pepstatin A (2mM), a specific inhibitor of aspartic proteases, showed inhibition of PrA residual activity (10%). Serine protease inhibitors, such as Pefabloc and PMSF (both at 5mM), caused inhibition of 20% and 12% of PrB, respectively. Pefabloc (5mM) and PMSF (1mM) caused strong inhibition of the residual activity of CpY, 12% and 3%, respectively. Chelating agents, such as EDTA (10mM) and 1–10 phenanthroline (10mM), did not exert a clear inhibitory effect, suggesting that these proteases are not metalloenzymes. Leupeptin (1mM) and E-64 (5mM) did not show significant inhibitory activity, which rules out the presence of cysteine groups in the catalytic domain of the proteases. The lack of inhibitory effect of bestatin excludes the presence of aminopeptidases (data not shown).
The presence of endogenous inhibitors of vacuolar proteases has been reported for S. cerevisiae.18 Since they could also exist in C. glabrata, the determination of enzymatic activity was conducted in conditions that inactivate these inhibitors. Accordingly and in order to evaluate PrA activity, this enzyme was preincubated at pH 5. PrB activity was assessed with the addition of SDS, and CPY activity by adding sodium deoxycholate.12
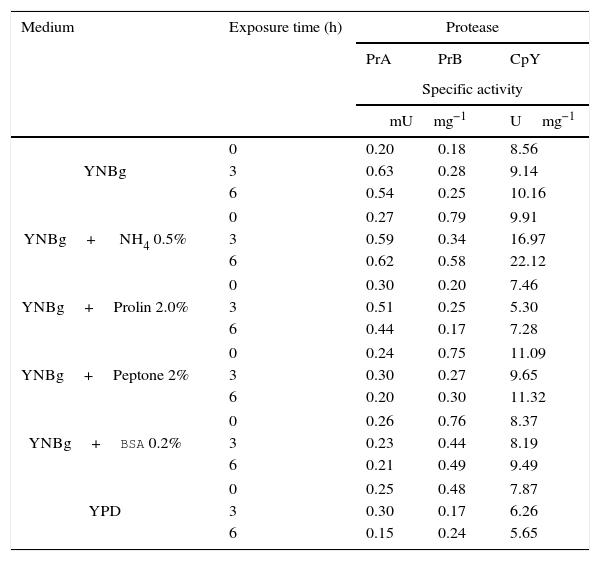
Proteolytic activity of putative vacuolar proteases under different physiological conditionsThe nitrogen source plays an important role in regulating the expression of protease-encoding genes in different organisms. Therefore, the expression can be measured at the level of enzyme activity or mRNA. To determine whether proteolytic activity is affected at the protein expression level by the nitrogen source, C. glabrata was cultured in YPD at early stationary phase (15h) and then tested in media with and without a nitrogen source. Accordingly, YNBg medium was supplemented with different nitrogen sources: ammonium, proline, peptone, and BSA. The level of protease activity was analyzed at 3 and 6h. The level of enzymatic specific activity of the three proteases studied showed little variation when making the transition from YPD to a different medium. Only the enzymatic activity of PrA and CpY increased (more than two-fold) in the presence of ammonium (basal value at 0h versus the value at 6h). Furthermore, with this nitrogen source PrA and CpY reached their maximum activity (Table 3).
Specificactivity of PrA, PrB and CpY from C. glabrata, determined from the cell free extract. The yeast was cultivated in YNB medium with different sources of nitrogen.
| Medium | Exposure time (h) | Protease | ||
|---|---|---|---|---|
| PrA | PrB | CpY | ||
| Specific activity | ||||
| mUmg−1 | Umg−1 | |||
| YNBg | 0 | 0.20 | 0.18 | 8.56 |
| 3 | 0.63 | 0.28 | 9.14 | |
| 6 | 0.54 | 0.25 | 10.16 | |
| YNBg+ NH4 0.5% | 0 | 0.27 | 0.79 | 9.91 |
| 3 | 0.59 | 0.34 | 16.97 | |
| 6 | 0.62 | 0.58 | 22.12 | |
| YNBg+Prolin 2.0% | 0 | 0.30 | 0.20 | 7.46 |
| 3 | 0.51 | 0.25 | 5.30 | |
| 6 | 0.44 | 0.17 | 7.28 | |
| YNBg+Peptone 2% | 0 | 0.24 | 0.75 | 11.09 |
| 3 | 0.30 | 0.27 | 9.65 | |
| 6 | 0.20 | 0.30 | 11.32 | |
| YNBg+BSA 0.2% | 0 | 0.26 | 0.76 | 8.37 |
| 3 | 0.23 | 0.44 | 8.19 | |
| 6 | 0.21 | 0.49 | 9.49 | |
| YPD | 0 | 0.25 | 0.48 | 7.87 |
| 3 | 0.30 | 0.17 | 6.26 | |
| 6 | 0.15 | 0.24 | 5.65 | |
PrA=Proteinase A; PrB=Proteinase B and CpY=Carboxypeptidase Y.
YNBg=medium containing yeast nitrogen base with 2% glucose.
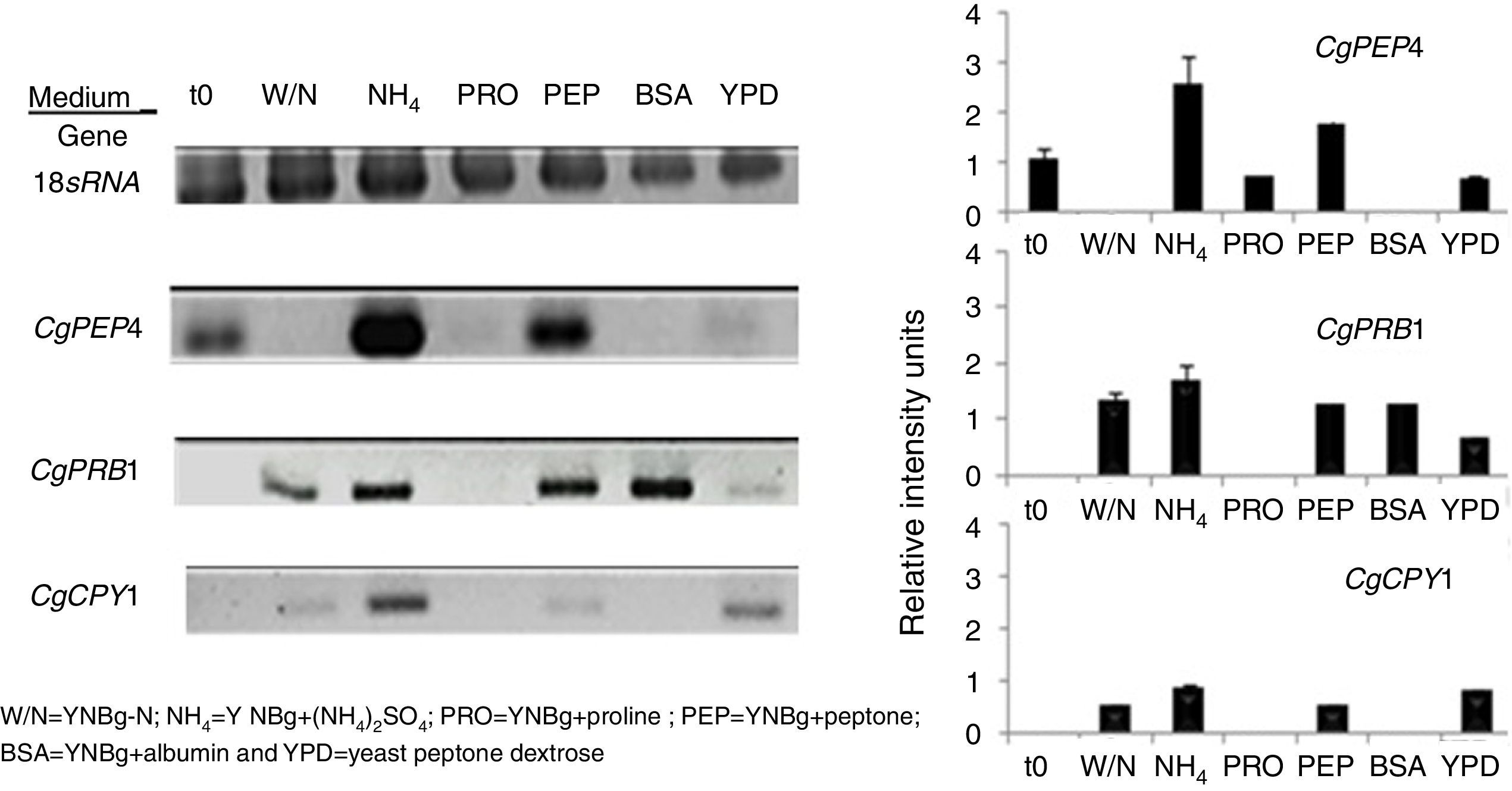
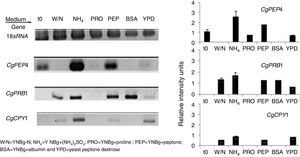
The regulation of expression of putative gene-encoding proteases in C. glabrata by distinct sources of nitrogen and under conditions of nutritional stress (nitrogen starvation) in several culture media at 6h was evaluated by RT-PCR (Fig. 3). Under the conditions studied, the CgPEP4, CgPRB1, and CgCPY1 genes were expressed in YNBg medium with ammonium as the nitrogen source. CgPRB1 was the only gene that was expressed with both, BSA as a nitrogen source and without a nitrogen source. Moreover, this gene also showed expression with ammonium and peptone; however, none of these genes were expressed in the YNBg medium with proline as the nitrogen source. The expression of genes in the YPD medium was not very evident, which is consistent with the levels of the specific activity found (Table 3).
DiscussionFungal vacuoles have been recognized as organelles that are involved in protein turnover, cellular homeostasis and membrane trafficking, and are essential for many of the attributes that define the functioning of fungi in their natural environments.42
C. glabrata displayed proteolytic activity similar to that found in some other yeasts, including S. cerevisiae, Schizosaccharomyces pombe, Kluyveromyces lactis, and Yarrowia lipolytica.9,10,24,37,38 Previous data revealed that the levels of PrA, PrB, and CpY proteases are dependent on the growth phase and composition of the nutrients in the growth media, e.g. higher levels of these proteases detected during early stationary phase in this study (data not shown). These results suggest that these proteases might be regulated by the source of nitrogen and carbon, as occurs with other fungi.9,10,37,38 Our findings based on the enzymatic activity of PrA, PrB and CpY proteases from the vacuolar fraction of C. glabrata displayed an increased activity suggesting their specific location in this organelle. The value of the enriched-activity yield for PrA is commensurate with the preparation of the pure vacuole as described in S. cerevisiae.33 Proteases that are considered canonical S. cerevisiae vacuolar enzymes could be used as vacuolar markers for C. glabrata.33
The regulation of the CgPEP4, CgPRB1 and CgCPY1 genes at the transcription level does not correspond completely to the levels of activity found for the enzymes coded by these genes. This may be due to post-translational processing of vacuolar proteases in this organelle, as described in S. cerevisiae or S. pombe.1,30,39 In C. albicans, it has been reported that the source of nitrogen and phase of growth influence the level of expression of vacuolar aspartic protease Apr1p (orthologous of PrA) and carboxypeptidase Cpy1p.3 On the other hand, it has been described that yeasts preferentially utilize glutamine, asparagine and ammonium as nitrogen sources, while proline and glutamate are poor sources of nitrogen.25 In the present study a differential expression was observed, according to the source of nitrogen employed in the three genes studied (CgPEP4, CgPRB1, and CgCPY); however, no expression of these three genes was found when they were tested in the presence of proline. Contrarily, an overexpression in C. albicans of dipeptidyl aminopeptidase, encoded by the CaSTE13 gene, was observed when using proline as the source of nitrogen.4 Intracellular proteolytic activity is very important for the protein turnover in S. cerevisiae, and may have the same function in C. glabrata.6
In this work, an acidic aspartyl proteinase (PrA) was detected in the C. glabrata vacuole, and the specific inhibitor Pepstatin A inhibited its activity. In S. cerevisiae, the ScPEP4 gene codes for an aspartyl protease, called proteinase A (Pep4) that is responsible for the sequential activation of many vacuolar hydrolases such as proteinase B and carboxypeptidase Y.13 The deletion of the ScPEP4 gene induces a reduction of the hydrolytic ability of the vacuole in S. cerevisiae. However, no detectable phenotypic consequences have been described in the C. albicans pep4Δ mutant.28 Hence, there may be significant differences between the proteolytic systems that operate in the vacuoles of C. albicans and the S. cerevisiae. Bioinformatics analysis in the C. glabrata genome showed only one putative CgPEP4 gene encoding for an acidic aspartyl proteinase located in a vacuole. The protein deduced from the CgPEP4 gene has a theoretical molecular mass similar to PrA of S. cerevisiae (45.4kDa for PrACg and 44.5kDa for PrASc). Moreover, a DD amino acid sequence of the catalytic domain confirms that the CgPEP4 gene codes for an aspartyl protease. The highest levels of mRNA expression of the CgPEP4 gene occurred in the presence of ammonium and peptone as sources of nitrogen. Contrarily, the absence of a nitrogen source did not cause an increase in the expression of this gene. The increase in CgPEP4 gene expression observed in culture media with ammonium or peptone as a nitrogen source may be due to the presence of GATA promoter sequences that exert physiological control over the catabolism of nitrogen (NCR) by two pathways, activation or repression.6 The CgPEP4 promoter sequence showed 21 putative binding sites for GATA proteins, while the ScPEP4 promoter sequence showed only one binding site for this protein. Coffman and Cooper suggest that global regulatory mechanisms are stimulated by nitrogen from distinct sources, including small molecules such as ammonium and macromolecules like peptone.6
Proteinase B activity was detected using HPA as a substrate, which resulted in a serine protease. In S. cerevisiae, proteinase B is involved in protein degradation in the vacuoles and is required for full protein degradation during sporulation.37 Therefore, PrB may be responsible for the activation of other vacuolar proteins in C. glabrata, and could participate in yeast turnover. Three ORFs coding for hypothetical PrB (Cg PRB1-3) were identified in the C. glabrata genome; meanwhile, in the S. cerevisiae genome only one ORF was found for this gene encoding for PrB. The proteins deduced from the CgPRB1-3 genes have a theoretical molecular mass from 50 to 63.6kDa that are homologous to the PrB protein of 69kDa in S. cerevisiae. It has been suggested that gene duplications in C. glabrata might be important adaptations for the success of this opportunistic pathogen. These duplications include the coding for glycosylphosphatidylinositol-linked aspartyl proteases by YPS genes, and for glycosylphosphatidylinositol-linked epithelial adhesins by EPA genes.7,31 The variation in the promoter sequences in each of the putative CgPRB genes suggests a differential expression between the three genes herein studied. The expression of the CgPRB1 gene would be regulated mainly by distinct sources of nitrogen, environmental changes, pH, and temperature. On the other hand, the CgPRB2 gene has a promoter sequence that responds to the beginning of cellular replication, while the CgPRB3 gene has a promoter sequence that is active during the cellular cycle. The duplication of CgPRB genes coding for PrB and the diversity of promoter sequences within each gene might provide C. glabrata with survival advantages under certain environmental conditions. Accordingly, it would be interesting to study the possible duplication of the CgPRB gene in C. glabrata31. Hence, the generation of a mutant in the CgPRB gene could clarify the role of the PrB protein in the life cycle of the yeast.
In this study, BSA was also employed as a nitrogen source in order to evaluate the expression of CgPEP4, CgPRB1, and CgCPY genes. Of the three genes herein studied, only CgPRB1 showed an increased expression in BSA. Although PrB is a protein with vacuolar location, it has been suggested that it participates in the protein turnover that helps the yeast survive. On the other hand, it has been demonstrated that other proteases, such as the aspartyl protease secreted by Sap2 of C. albicans and C. dubliniensis, induce the expression of PrB in the presence of BSA.20,25 The activity of the PrB protease may support the adaptation and survival of C. glabrata in the host, especially during systemic infections.35
In the present study, results performed with the CpY protease of C. glabrata displayed a serine protease activity in the soluble fraction. Besides, theoretical analysis of CpY protease showed a molecular mass of 57kDa, while the homologue enzyme of S. cerevisiae has a molecular mass of 60kDa. Upregulation of CgCPY1 gene expression and activity occurred in the YNBg-ammonium medium. In C. albicans, the determination of CaCPY1 gene expression under nitrogen-limited conditions revealed that this gene was regulated at both the transcriptional and translational levels.3 In S. cerevisiae, carboxypeptidase activity is involved in low-nitrogen conditions, sporulation, process of killing toxins, and non-specific protein degradation in the vacuole; in addition, carboxypeptidase Y is also considered a vacuolar marker enzyme.33
C. glabrata can survive in nutrients-poor conditions (e.g. on the surface of epithelial cells and inert surfaces like catheters where the yeast can form biofilms).36 Another characteristic of this yeast is its capacity to survive in macrophages, which implies that it can endure the respiratory burst.35 Roetzer et al. demonstrated the formation of peroxisomes in yeasts and the activation of autophagy involved in their capacity of intracellular survival,32 although few studies have been conducted in this regard. There have been reports about the preponderant role of the yeast vacuole and its proteases during autophagy, assisting in the degradation and protein turnover under stressful conditions.16,43 Consequently, the study of vacuolar proteases herein described, as well as other proteases such as aminopeptidases, should allow researchers to elucidate the role played in the survival and proliferation of C. glabrata during the process of host infection. These results establish the basis for the biochemical and molecular study of vacuolar proteases in C. glabrata. The role of vacuole-mediated proteolysis in this yeast may be explored by obtaining the corresponding mutants and studying host-pathogen interactions.
Conflict of interestThe authors declare that they have no conflict of interests in relation to any of the techniques or materials used in this study.
We thank Bruce Allan Larsen for reviewing the use of English in this manuscript. We are grateful to Bernard Dujon for donating the strains; BPO and YBC were fellows from CONACyT and BPO was PIFI-IPN; LVT and CHHR received support from COFAA-IPN and EDI-IPN. Grants from CONACyT (133695, 14299, 69984, 182651, and 133451), (SIP-IPN-2015-0981 and 2014-1333), and Federal Funds HIM/2011/012 are also acknowledged.